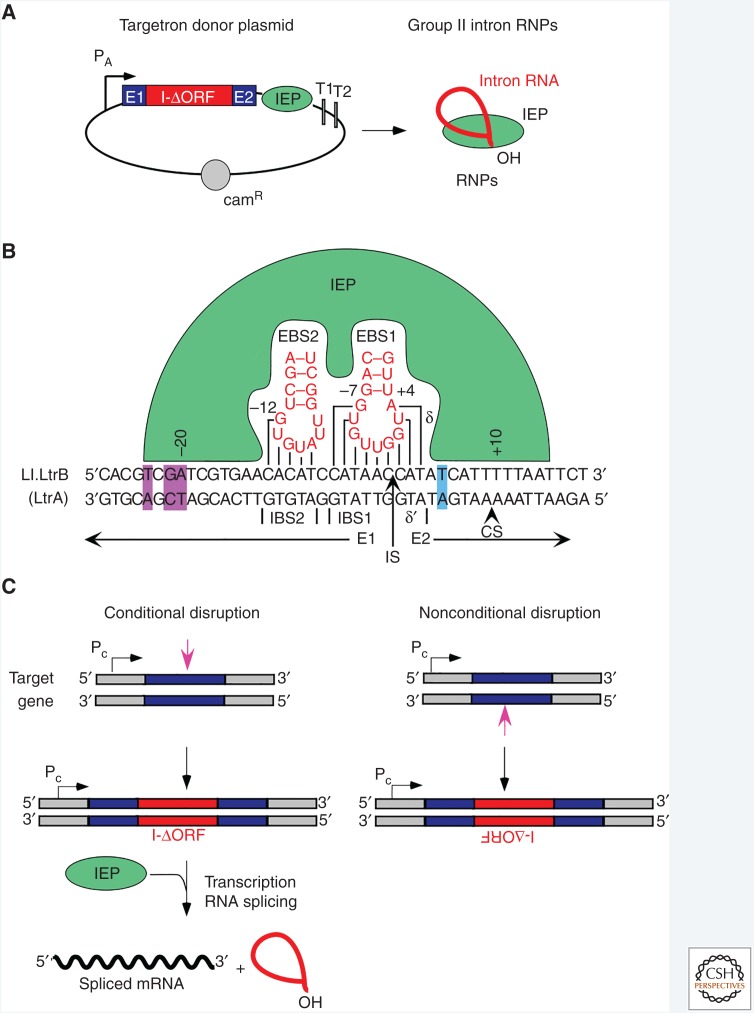
Figure 4.
Group II introns as targetrons for bacterial gene targeting. (A) Targetron donor plasmid. The plasmid has a strong and preferably inducible active promoter (PA) to express a targetron cassette consisting of a group II intron ribozyme with the reverse transcriptase (RT) open reading frame (ORF) deleted (I-ΔORF) flanked by short 5′ and 3′ exons (E1 and E2, respectively) and followed by an ORF encoding the intron-encoded protein (IEP). The IEP splices the group II intron RNA and remains bound to the excised intron lariat RNA in an RNP that can be preprogrammed to insert into desired target sites. (B) DNA target site interactions for the Ll.LtrB group IIA intron used for gene targeting involve base-pairing of EBS1, EBS2, and δ sequences in DI with IBS1, IBS2, and δ′ sequences flanking the intron insertion site (IS). The IEP recognizes specific nucleotides (purple) in the distal 5′-exon region upstream of IBS2 and promotes local DNA melting, enabling base-pairing of the intron RNA to the DNA target sequence. Additional interactions between the IEP and nucleotides in the 3′ exon (blue) are required for bottom-strand cleavage by the EN domain to generate the primer for reverse transcription of the intron RNA, leading to intron insertion via the retrohoming pathway. CS, bottom-strand cleavage site. (C) Use of targetrons for conditional or nonconditional disruptions. Targetrons can be made to insert in either orientation relative to transcription of the target gene (dark blue) by targeting a sequence (magenta arrow) in the top or bottom strand. A targetron that inserts in the antisense orientation yields an unconditional disruption, whereas a targetron that inserts in the sense orientation can yield a conditional disruption by linking splicing of the intron from the pre-mRNA to expression of the IEP from a separate construct. Pc, host chromosomal gene promoter. (Adapted from Enyeart et al. 2014.)