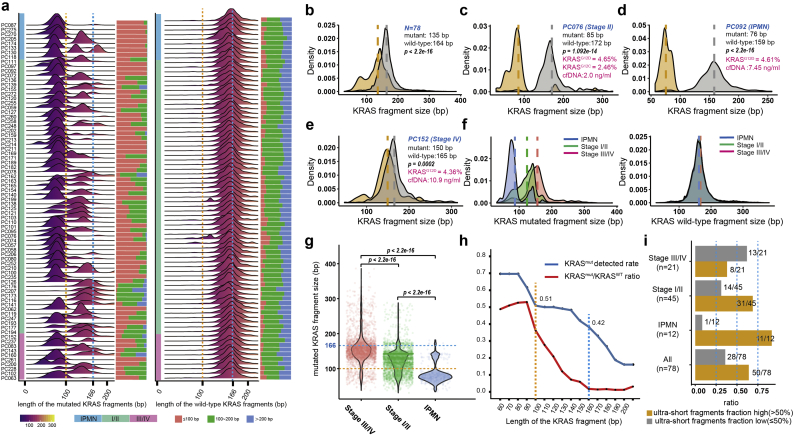
Fig. 3.
Ultra-short fragments containing the KRAS hotspot alleles in patients with pancreatic cancer. (a) Comprehensive view of fragment lengths bearing KRAS mutated and wild-type alleles derived from cell-free DNA deep-sequencing in patients with pancreatic cancer (n = 78). (b) The fragment length of cfDNA bearing mutant KRAS alleles tended to be significantly shorter compared with DNA fragments bearing wild-type allele by densitometry in pancreatic cancer (n = 78). The fragments bearing KRAS mutant alleles tended to be enriched in the region below 100 bp in patients (c) PC076 (stage II) and (d) PC092 (IPMN). (e) The distribution of KRAS mutated fragments included a large proportion of overlapping fragment sizes with wild-type KRAS fragments in advanced patients (PC152). (f) There was considerable discrepancy across IPMN (n = 10), early-stage (n = 42) and advanced (n = 26) pancreatic cancer in terms of the length of KRAS mutated fragments by densitometry. (g) Violin plots representing the length of KRAS mutated fragments across different disease stages. (h) The ability of library preparation to enrich shorter fragments has a great impact on the detection of KRAS hotspot mutations in plasma (blue line), and the mutated-to-wild-type fraction of fragments bearing KRAS alleles reached the highest in the interval of 60–100 bp and declined sharply thereafter (red line). (i) Ultra-short fragments (<100 bp) accounted for a quite high proportion of total KRAS mutated fragments in a substantial group of patients with pancreatic cancer. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)