Table 3.
Top 20 immunogenic antigens in patients with AMVR of 857 candidate antigens overexpressed in microvascular ECs
| Gene ENS | Symbol | Δ Expression in Micro-ECs versus Macro-ECs | Protein Locus | Description | Frequency in Stable Patients, % | Frequency in Patients with AMVR, % | Intensity in Stable Patientsa | Intensity in Patients with AMVRa | Overall Scoreb | P Value |
|---|---|---|---|---|---|---|---|---|---|---|
| ENSG00000162078 | ZG16B | 148.5 | NM_145252.1 | Zymogen granule protein 16B | 33.33 | 90.91 | 2.11 | 2.92 | 87.2 | <0.001 |
| ENSG00000163431 | LMOD1 | 144.8 | BC001755.1 | Leiomodin 1 | 25.00 | 68.18 | 1.04 | 1.99 | 60.4 | 0.01 |
| ENSG00000107779 | BMPR1A | 1782.7 | NM_004329 | Bone morphogenetic protein receptor, type IA | 16.67 | 72.73 | 0.87 | 1.03 | 57.4 | 0.001 |
| ENSG00000197971 | MBP | 4251.9 | NM_001025100.1 | Myelin basic protein | 25.00 | 63.64 | 1.11 | 2.16 | 56.4 | 0.03 |
| ENSG00000169188 | APEX2 | 23.7 | NM_014481.2 | APEX nuclease 2 | 25.00 | 77.27 | 0.93 | 1.09 | 55.0 | 0.01 |
| ENSG00000106789 | CORO2A | 433 | NM_052820.1 | Coronin, actin binding protein, 2A | 33.33 | 63.64 | 1.01 | 2.34 | 51.1 | 0.18 |
| ENSG00000183287 | CCBE1 | 15,174.7 | BC046645.1 | Collagen and calcium binding EGF domains 1 | 8.33 | 45.46 | 0.60 | 1.59 | 46.0 | 0.06 |
| ENSG00000145242 | EPHA5 | 1609.2 | PV3359 | EPH receptor A5 | 25.00 | 63.64 | 0.90 | 1.23 | 44.0 | 0.08 |
| ENSG00000106829 | TLE4 | 1281.9 | BC059405.1 | Transducin-like enhancer of split 4 | 33.33 | 68.18 | 2.05 | 1.99 | 43.5 | 0.05 |
| ENSG00000142459 | EVI5L | 1270.2 | NM_145245.1 | Ecotropic viral integration site 5-like | 25.00 | 59.09 | 0.84 | 1.33 | 41.4 | 0.12 |
| ENSG00000107679 | PLEKHA1 | 2978.9 | NM_001001974.1 | Pleckstrin homology domain containing, family A1 | 16.67 | 45.46 | 0.69 | 1.71 | 39.7 | 0.06 |
| ENSG00000198959 | TGM2 | 28,594.6 | BC003551.1 | Transglutaminase 2 | 8.33 | 45.46 | 0.68 | 0.96 | 37.6 | 0.06 |
| ENSG00000082805 | ERC1 | 2673.4 | PV3626 | ELKS/RAB6-interacting/CAST family member 1 | 8.33 | 31.82 | 0.39 | 2.31 | 36.0 | 0.07 |
| ENSG00000198081 | ZBTB14 | 285.7 | NM_003409.2 | Zinc finger and BTB domain containing 14 | 33.33 | 63.64 | 0.94 | 1.25 | 36.0 | 0.18 |
| ENSG00000128872 | TMOD2 | 560.4 | BC036184.1 | Tropomodulin 2 | 16.6 | 50.00 | 0.85 | 1.09 | 35.6 | 0.04 |
| ENSG00000168175 | MAPK1IP1L | 3083.3 | NM_144578.1 | Mitogen-activated protein kinase 1 interacting protein 1-like | 8.33 | 40.91 | 0.60 | 1.12 | 35.5 | 0.02 |
| ENSG00000112561 | TFEB | 171.1 | NM_007162.1 | Transcription factor EB | 8.33 | 40.91 | 0.62 | 0.99 | 33.7 | 0.02 |
| ENSG00000123836 | PFKFB2 | 596.2 | NM_006212.1 | 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 | 41.67 | 68.18 | 1.09 | 1.40 | 33.4 | 0.12 |
| ENSG00000106123 | EPHB6 | 119.4 | NM_004445.1 | EPH receptor B6 | 8.33 | 36.36 | 0.69 | 1.14 | 30.6 | 0.15 |
| ENSG00000240694 | PNMA2 | 684.4 | XM_376764.2 | Paraneoplastic ma antigen 2 | 8.33 | 36.36 | 0.65 | 1.10 | 30.3 | 0.04 |
ENS, Ensembl Gene ID.
Intensity represents the average ratio of observed reactivity exceeding the cut-off for sera from stable patients and patients with AMVR.
The score was calculated using the equation 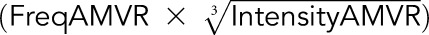 −
−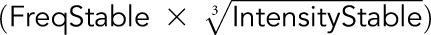 described by Gnjatic et al.7
described by Gnjatic et al.7
