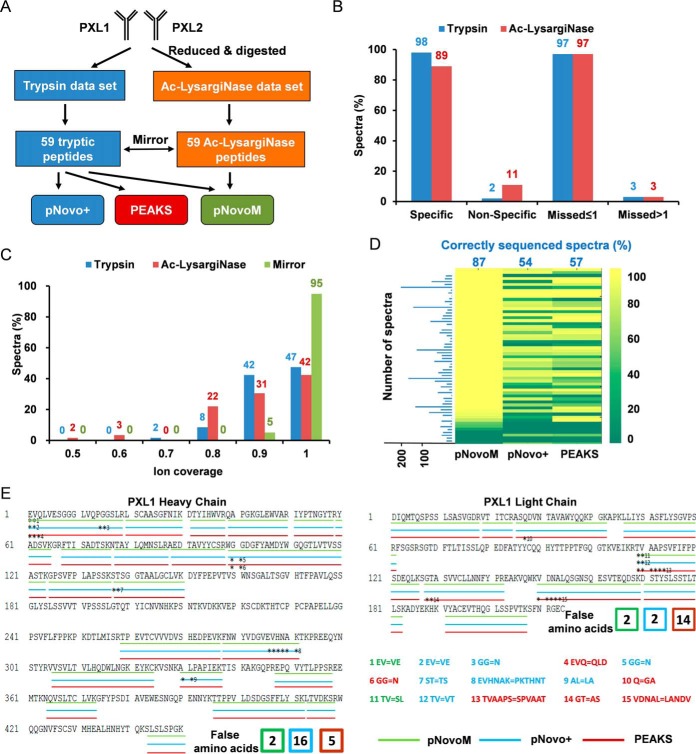
Fig. 3.
Superior precision de novo sequencing of monoclonal antibody PXL1 based on the mirror protease strategy. A, Workflow of the analysis of two antibody data sets (PXL1 and PXL2) generated from trypsin- and Ac-LysargiNase-digested samples. B, Comparison of the specificity and miss cleavage of the spectra sequenced from trypsin or Ac-LysargiNase-digested antibody PXL1. C, The distribution of ion coverage of trypsin, Ac-LysargiNase and mirror spectra. D, Identification rates of the spectra for each peptide from pNovoM, pNovo+ and PEAKS. The blue numbers indicate the average percentage of the correctly sequenced mirror spectrum pairs, and the blue bar chart denotes the number of mirror spectrum pairs for each mirror peptide pair. E, The peptides sequenced by three algorithms on the heavy and light chains of PXL1. The three different color lines were the results of pNovoM (green), pNovo+ (blue) and PEAKS (red). The asterisks denote incorrect residues. The false amino acids matching was listed as the number showed.