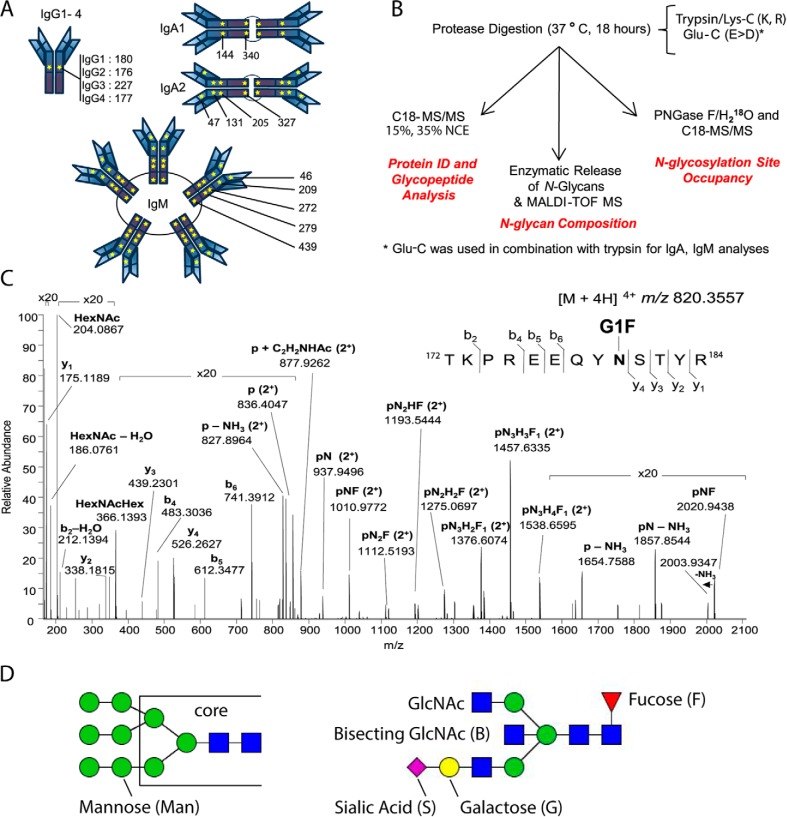
Fig. 1.
N-Glycosylation Sequons in the Immunoglobulin Fc Region Are Present in All Antibody Isotypes and Subclasses. A, Schematic of immunoglobulin N-glycosylation sequons. Stars represent the locations of N-glycosylation sequons within the Fc regions of IgG1–4, IgA1–2 and IgM. The location of the asparagine (N) within each sequon is numbered from the N terminus of the heavy chain constant region, and thus relates to the constant chain only. B, Schematic of the analytical workflow used in this study. nLC-MS/MS that employed C18 as the solid phase; stepped collision energy (15%, 35%) was used to observe fragments arising from cleavage of carbohydrate glycosidic linkages and peptide backbone fragments, respectively. Enzymatic release of N-glycans in H218O was achieved via the use of PNGase F. The PNGase release was also used to introduce 18O into formerly glycosylated asparagine residues, to estimate glycosylation site occupancy. C, Higher-energy collisional dissociation (HCD) spectrum of IgG1 N-glycopeptide 172TKPREEQYNSTYR184 + HexNAc4Hex4Fuc1 ([M + 4H]4+ m/z 820.3551), observed with 0.63 ppm error. Peptide b- and y-ions arising from breakage of the peptide bond and fragments arising from cleavage of glycosidic linkages, are labeled. D, Nomenclature for high mannose (left) and complex (right) N-linked glycans. High mannose and complex N-linked glycans share a common tri-mannosyl-chitobiose core (core). High mannose glycans are named by listing the total number of mannose (Man) residues. Man9, with two N-acetylglucosamine (GlcNAc) residues and nine mannose residues, is shown (left). A complex glycan consists minimally of the tri-mannosyl-chitobiose core with a single GlcNAc linked to each branch, and these residues are not listed. Monosaccharides beyond this basic structure are listed. G1S1FB (shown, right) is extended on one branch with galactose (G) and sialic acid (S), modified at the core with a fucose (F), and bisected with an N-acetylglucosamine (B).