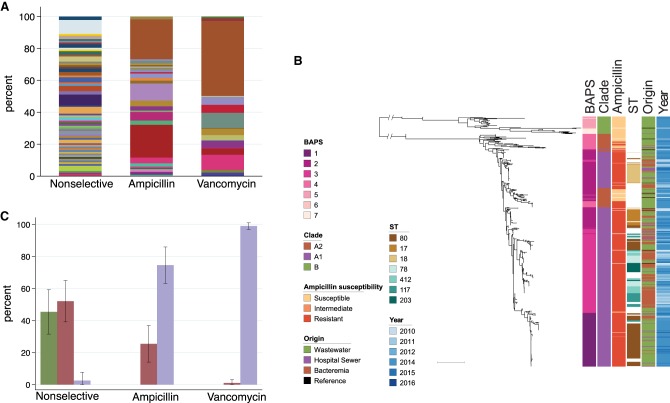
Figure 2.
Genetic characterization of E. faecium isolates from wastewater and bloodstream infections. (A) MLST sequence types of 383 E. faecium isolates recovered from wastewater according to antibiotic selection during isolation. Nonselective: no selection for antibiotic resistance; ampicillin: ampicillin-resistance selection; vancomycin: vancomycin-resistance selection. (B) Midpoint rooted maximum-likelihood tree based on SNPs in 1336 core genes of 620 E. faecium (383 wastewater, 40 hospital sewage, 187 from bloodstream, and 10 reference isolates) labeled by hierarchical Bayesian cluster (1–7), clade (B, A2, and A1), ampicillin-susceptibility, commonest STs, origin, and year of isolation. Scale bar, ∼10,000 SNPs. (C) Proportion of each E. faecium clade recovered from wastewater according to antibiotic selection during isolation. A single isolate of clade A2 recovered on the vancomycin selective plate was vancomycin-susceptible. Error bars, standard deviation.