Figure 1.
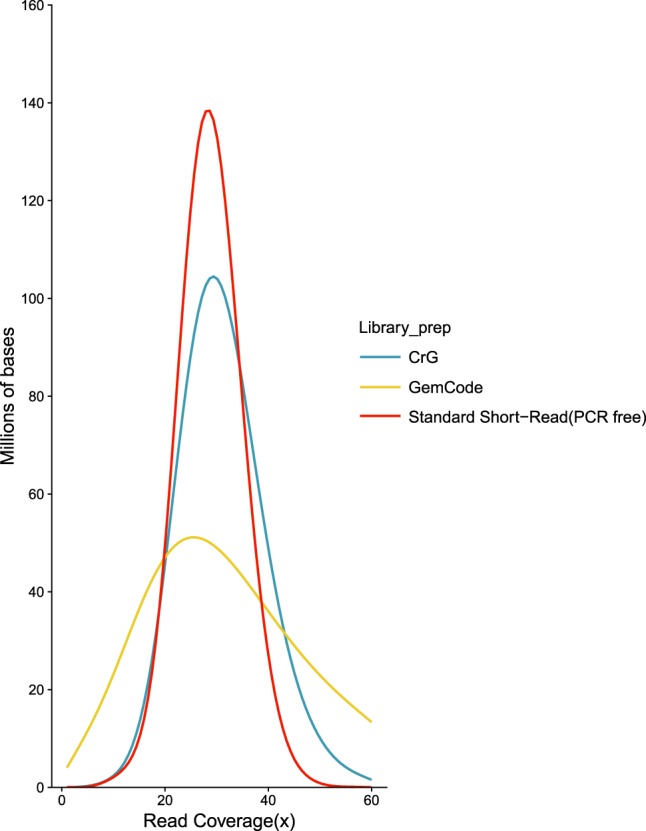
Coverage evenness. Distribution of read coverage for the entire human genome (GRCh37). Comparisons between 10x Genomics Chromium Genome (CrG), 10x Genomics GemCode (GemCode), and Illumina TruSeq PCR-free standard short-read NGS library preparations (Standard Short-Read [PCR-Free]). Sequencing was performed in an effort to match coverage (Methods). Note the shift of the CrG curve to the right, showing the improved coverage of Chromium versus GemCode. The x-axis represents the fold read coverage across the genome, and the y-axis represents the total number of bases covered at any given read depth.
