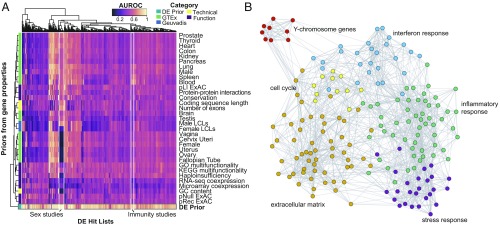
Fig. 3.
Multiple independent gene processes underlie the high performance of the DE prior. (A) Heatmap of AUROCs for each gene property-related prior (rows) and each study in the compendium (columns). Row colors indicate the gene property type. No single property performs as well as the DE prior. However, a few studies are clearly predicted by a number of features, including sex-related studies and immunity-related studies (both highlighted with white boxes). These groups are well predicted by expression level in males and expression level in blood, respectively. (B) Network visualization of genes in the top 1% of the DE prior. Each circle represents a gene, and edges connect all genes that are nearest neighbors based on their coexpression in an aggregate network. Genes are colored by cluster and labeled by GO enrichment. We find that genes in the top 1% of the prior show strong coexpression patterns, representing distinct functional modules that are commonly repurposed across many different conditions. ExAC, Exome Aggregation Consortium; GO, Gene Ontology; GTEx, Genotype-Tissue Expression project; KEGG, Kyoto Encyclopedia of Genes and Genomes; LCLs, lymphoblastoid cell lines; pLI, probability of intolerance to a single loss-of-function variant; pNull, probability of tolerance to loss-of-function variation; pRec, probability of intolerance of two loss-of-function variants.