Significance
There is great interest in spreading beneficial traits throughout wild populations in self-sustaining ways. Here, we describe a synthetic selfish genetic element, CleaveR [Cleave and Rescue (ClvR)], that is simple to build and can spread a linked gene to high frequency in populations. ClvR is composed of two components. The first, germline-expressed Cas9 and guide RNAs (gRNAs), cleave and disrupt versions of an essential gene located elsewhere in the genome. The second, a version of the essential gene resistant to cleavage, provides essential gene function. ClvR spreads by creating conditions in which progeny lacking ClvR die because they have no functional copies of the essential gene. In contrast, those who inherit ClvR survive, resulting in an increase in ClvR frequency.
Keywords: gene drive, Cas9, population replacement, selfish genetic element
Abstract
There is great interest in being able to spread beneficial traits throughout wild populations in ways that are self-sustaining. Here, we describe a chromosomal selfish genetic element, CleaveR [Cleave and Rescue (ClvR)], able to achieve this goal. ClvR comprises two linked chromosomal components. One, germline-expressed Cas9 and guide RNAs (gRNAs)—the Cleaver—cleaves and thereby disrupts endogenous copies of a gene whose product is essential. The other, a recoded version of the essential gene resistant to cleavage and gene conversion with cleaved copies—the Rescue—provides essential gene function. ClvR enhances its transmission, and that of linked genes, by creating conditions in which progeny lacking ClvR die because they have no functional copies of the essential gene. In contrast, those who inherit ClvR survive, resulting in an increase in ClvR frequency. ClvR is predicted to spread to fixation under diverse conditions. To test these predictions, we generated a ClvR element in Drosophila melanogaster. ClvRtko is located on chromosome 3 and uses Cas9 and four gRNAs to disrupt melanogaster technical knockout (tko), an X-linked essential gene. Rescue activity is provided by tko from Drosophila virilis. ClvRtko results in germline and maternal carryover-dependent inactivation of melanogaster tko (>99% per generation); lethality caused by this loss is rescued by the virilis transgene; ClvRtko activities are robust to genetic diversity in strains from five continents; and uncleavable but functional melanogaster tko alleles were not observed. Finally, ClvRtko spreads to transgene fixation. The simplicity of ClvR suggests it may be useful for altering populations in diverse species.
Gene drive occurs when particular alleles are transmitted to viable, fertile progeny at rates greater than those of competing allelic variants. Strategies for altering the genetics of populations that incorporate some level of drive to enhance the spread of linked transgenes, but that are not self-sustaining, have been proposed but not yet implemented (1–3). A number of approaches to spreading traits through populations (population replacement/alteration) in ways that are self-sustaining, by linking them with genetic elements that mediate drive, have also been proposed (4–16). Much recent interest has focused on approaches to population alteration that utilize engineered site-specific nucleases that function as homing endonuclease genes (HEGs) (17). A HEG encodes a site-specific nuclease that is inserted within its chromosomal recognition sequence. This prevents cleavage of the homolog within which it resides. If, in a heterozygote, the wild-type allele is cut and homologous recombination (HR) is used as the repair pathway with the HEG-bearing chromosome as the repair template, the HEG heterozygote can be converted into a homozygote (also known as homing), thereby increasing HEG copy number. There is particular interest in HEGs created using the CRISPR/Cas9 endonuclease system, in which the Cas9 endonuclease is targeted to specific sequences through association with one or more independently expressed guide RNAs (gRNAs) (18). Target sequence limitations with Cas9 are modest, and thus Cas9 in conjunction with one or more gRNAs can be used to cleave a gene at multiple positions, making these reagents ideal tools for HEG engineering. Population alteration using HEGs can in principle be achieved in several ways (17, 19). However, all require homing, which requires that cleavage be followed by repair and copying of the intact HEG through high-fidelity HR. While important progress has been made, sustained alteration of a population [as opposed to suppression (20)] to transgene-bearing genotype fixation with a synthetic HEG into artificial or naturally occurring sites remains to be achieved (21–30).
A number of other approaches to bringing about gene drive take as their starting point naturally occurring, chromosomally located, selfish genetic elements whose mechanism of spread does not involve homing (4, 6, 31). Many of these elements can be represented as consisting of a tightly linked pair of genes encoding a trans-acting toxin and a cis-acting antidote that neutralizes toxin expression and/or activity (TA systems) (4). The general idea is often that toxin expression or activity is repressed in cells that carry the TA pair because they also express the antidote, allowing survival. However, when such a system is present in an organism, those gametes, progeny, or daughter cells that fail to inherit the TA system die because the toxin or effects of toxin activity remain present, while the cis-acting antidote is absent: a phenomenon known as postsegregational killing. Examples of such systems where some molecular information is available include the maternal-effect selfish genetic element Medea in Tribolium (32, 33), the sup-35/pha-1 maternal-effect selfish genetic element in Caenorhabditis elegans (34), the peel-zeel paternal-effect selfish genetic element in C. elegans (35), and the wtf gamete/spore killers in yeast (36, 37). Synthetic Medea elements generated in Drosophila use a similar logic, but with the toxin simply being a maternally expressed miRNA (the toxin) that results in maternal loss of a product normally deposited into the embryo that is essential for early embryo development (the consequences of toxin expression). The antidote is a transgene that results in early embryonic expression of a recoded version of this same gene that is resistant to miRNA silencing (the antidote), thereby providing essential gene function in a just-in-time fashion (6, 38, 39). Finally, prokaryotes also contain a number of tightly linked toxin–antidote clusters (including but not limited to type II restriction enzymes and their cognate methyltransferases). While many of these play important roles in cell physiology and defense, there are also multiple lines of evidence showing that some of them act in a selfish manner to increase their representation within populations through postsegregational killing of those that fail to inherit them (and thus the antidote) at cell division as a result of inefficient partitioning, or when in competition with other similar units (plasmids of the same incompatibility group/replicon) that lack them (40, 41). Based on these behaviors bacterial TA systems are sometimes known as addiction modules: the components they encode are fundamentally nonessential (as with the eukaryotic TA systems described above), but once they are acquired, they cannot easily be lost without causing death of the host cell.
The components of naturally occurring TA systems could in principle be adapted to bring about gene drive in other species of interest. While the locus that contains Tribolium Medea has been sequenced, the molecular nature of TA components that account for its behavior remains unknown. Toxin and antidotes associated with C. elegans maternal- and paternal-effect selfish genetic elements are known (35, 34), as are those associated with gamete/spore killing in yeast (36, 37). However, in these latter cases, it is unclear whether the mechanisms of action and any associated gene regulation required for selfish behavior can be transferred across species. Implementation of synthetic Medea was successful in Drosophila, but this relied on detailed knowledge of the molecular genetics that underlie maternal and early zygotic control of embryogenesis. Efforts to translate Medea to species other than the closely related Drosophila suzukii (39) have not yet succeeded. Toxins and antidotes from prokaryotes are well understood at a mechanistic level, and are often likely to be active in eukaryotic systems since many of them target highly conserved processes, such as translation, or promote the degradation of RNA or DNA (40–42). However, the use of these or other gain-of-function toxins and antidotes requires careful titration of the place and time they are transcribed and translated. Achieving such control, as with synthetic Medea elements, is likely to require a deep species-specific toolbox of information and reagents, including knowledge of details of development, promoters, and regulators of translation and degradation during key stages of development, such as the maternal-zygotic transition. In sum, while existing TA systems are attractive to consider as a starting point for development of new gene drive systems—since they bring about drive in nature—the available tools do not yet provide a straightforward and general approach to building TA-based chromosomal gene drive methods in diverse species.
Here, we report the creation of a TA-based chromosomally located selfish genetic element whose components are simple and interchangeable, and likely to be generally available across species. Our starting point is the fact that site-specific alteration of DNA in the germline, mediated by Cas9 and gRNAs or other site-specific nucleases, followed by error-prone repair or creation of larger deletions, can be used most simply to disrupt the function of a gene, in our case an essential gene. Site-specific base editing enzymes (43) can be employed toward a similar end. Here, we focus on site-specific nucleases. Novel versions of essential genes that share limited or no nucleotide sequence similarity with the endogenous version, and are thus uncleavable, can rescue the viability and fertility of individuals that otherwise carry only loss-of-function (LOF) versions of the essential gene (44–46). Recombination and gene conversion can occur between a cleaved locus and an uncleaved counterpart located elsewhere in the genome to which it has sequence similarity (47), and this could lead to the creation of functional, cleavage-resistant alleles at the endogenous essential gene locus. Reducing or eliminating sequence similarity between the cleaved version of the essential gene and an uncleavable rescuing version can prevent such events (48). Finally, in the case of diploids, for many essential genes (haplosufficient recessive lethal or sterile), heterozygotes for a LOF allele are, at least to a first approximation, fit (49–51).
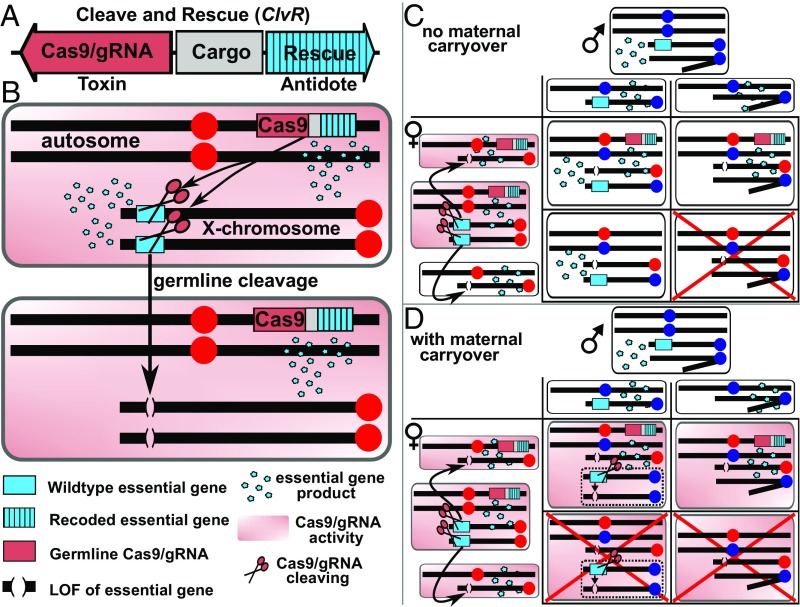
Under the above conditions, a cassette that includes germline-expressed Cas9 and gRNAs, designed to cleave in trans and thereby disrupt any endogenous wild-type copies of an essential gene, and a recoded version of the essential gene resistant to cleavage and recombination or gene conversion with cleaved versions of the wild-type allele, and therefore able to rescue those who carry it in cis, behaves as a selfish genetic element, which we refer to as CleaveR [Cleave and Rescue (ClvR)] (Fig. 1A). The toxin, Cas9 and gRNAs, works in trans by creating a permanent, potentially lethal change to the host genome wherever the targeted locus is located. However, this lethality only manifests itself in those who fail to inherit ClvR and its cis-acting antidote, the Rescue transgene. In contrast, those who inherit ClvR and the Rescue transgene contained within it survive, resulting in an increase in the frequency of individuals with ClvR-bearing chromosomes compared with those carrying non–ClvR-bearing counterparts. (Fig. 1, and other examples in SI Appendix, Figs. S1 and S2). This represents a form of postsegregational killing and leads cells, organisms, and populations to become dependent on (addicted to) the ClvR-encoded Rescue transgene (the antidote) for their survival. An analogy can be drawn with one strategy used to force the maintenance of a costly, nonessential plasmid in the absence of antibiotic selection. This involves locating an unconditionally essential gene (normally chromosomal) on the plasmid in cells that otherwise lack a functional copy of the essential gene (52). A ClvR element simply has the added feature that it provides the mechanism by which the endogenous version of the essential gene is inactivated in addition to the mechanism promoting survival in its absence. In Results and Discussion, we consider the specific case of ClvR behavior in a diploid animal, Drosophila melanogaster, as a model for other species such as mosquitoes, for which there has long been interest in the idea of altering wild populations so that they are unable to transmit diseases such as dengue, yellow fever, chikungunya, or malaria.
Fig. 1.
Basic structure of a ClvR element, and its behavior in a diploid, with and without maternal carryover. (A) Components of a ClvR element. (B) Behavior of ClvR as implemented for a ClvR on an autosome and an essential gene located on X chromosome. The long thick horizontal black bar represents a chromosome with ClvR on the right arm of an autosome (see experiments below for an experimental implementation), while the shorter horizontal black bar represents an X chromosome carrying an essential gene. The identity of genes, alleles, and protein and RNA products are indicated. Arrows are drawn from a wild-type allele of the essential gene to the cleaved product resulting from Cas9 activity. (C) Results of a cross between a heterozygous ClvR-bearing female and a wild-type male, in the absence of maternal carryover of Cas9/gRNA complexes. Arrows indicate conversion from wild-type to LOF allele. (D) Same cross as in C, but with maternal carryover of Cas9/gRNAs sufficient to convert wild-type alleles of the essential gene inherited from the father into LOF alleles. The dashed boxes highlight the paternal X chromosome before and after cleavage and creation of a LOF allele. Arrows indicate conversion from wild-type to LOF allele. Large red Xs indicate offspring that die because they lack any source of essential gene function. The color of the centromere (large circle) indicates whether the chromosome was inherited from a female (red) or male (blue) parent. The Y chromosome is shown as a short horizontal black bar with an angled segment, and a blue centromere.
Results and Discussion
ClvR and the locus it targets for inactivation can be located on the same chromosome or on different chromosomes. The specific relationship is not important for gene drive since cleavage occurs in trans, wherever the target gene is located, while rescue only occurs in cis, in those who inherit ClvR. ClvR behavior is illustrated in Fig. 1 B–D for the case in which ClvR is located on an autosome and the haplosufficient essential gene targeted for cleavage is located on the X chromosome (see below and Figs. 2–5 for related experiments). Cleavage by Cas9 followed by inaccurate repair creates LOF alleles of the essential gene in the adult female germline (Fig. 1B). Diploid germ cells survive because they carry a copy of ClvR, which includes the recoded Rescue. In animals, haploid gametes lacking ClvR and a functional copy of the essential gene (e.g., some female gametes in Fig. 1 C and D) will generally survive and be functional because essential gene products utilized during the haploid stage are expressed during the diploid stage and shared between the products of meiosis (53–55). However, in other organisms in which extensive transcription occurs during the haploid stage (e.g., plants and fungi), gametes lacking ClvR will be lost if transcription of the targeted essential gene is required during the haploid phase for gamete survival or function (SI Appendix, Fig. S1).
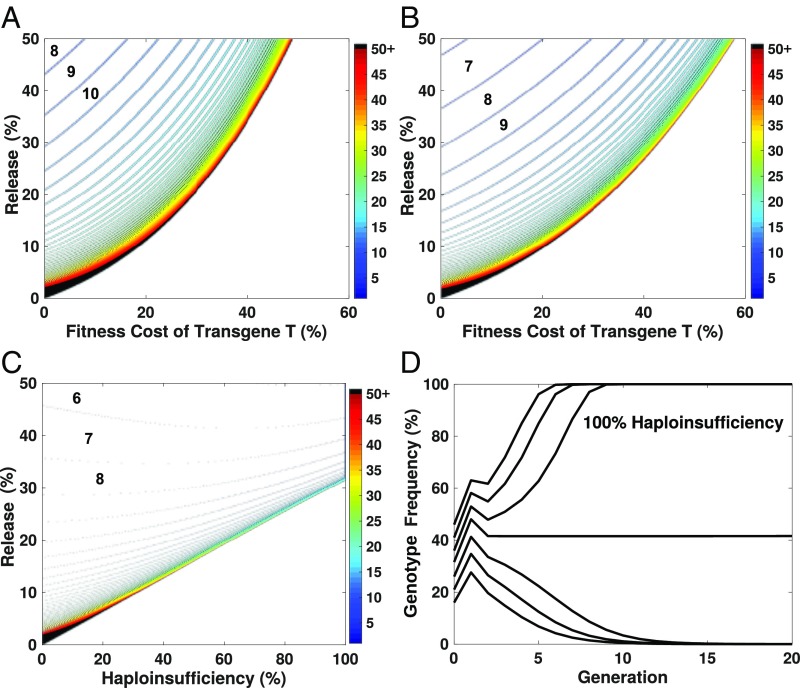
Fig. 2.
Population genetic behavior of ClvR when targeting a haplosufficient (A and B) or haploinsufficient (C and D) essential gene. (A and B) A discrete generation, deterministic population frequency model of ClvR spread in which cleavage occurs in the male and female germline; ClvR located on an autosome and the essential gene is located on the X (see data in Figs. 3 and 5) through a single panmictic population, for varying initial release percentages and fitness costs, without (A) or with (B) maternal carryover-dependent cleavage. The heatmap indicates the number of generations required for the ClvR-bearing genotype to approach fixation (i.e., >99% of the total population). (C) Heatmap showing the number of generations required for the ClvR-bearing genotype to reach fixation (<99% ClvR-bearing) for different initial release percentages and haploinsufficient fitness costs (100% = haplolethal), for an autosomal version of ClvR targeting a second unlinked autosomal locus, with maternal carryover. (D) Individuals traces showing the fate of a ClvR from (C) targeting a haplolethal gene, for different release percentages. The horizontal line represents an approximation of the unstable equilibrium frequency (∼31.5%; genotype frequencies do not change significantly over 20 generations). Genotype frequencies greater than equilibrium, 36%, 41%, and 46%; those below, 26%, 21%, and 16%. Note that the term “Release %” for all heatmaps refers to the percentage of homozygous transgenic males compared with wild-type males and females after a release has occurred (e.g., a 40% release means that 40% of the population is ClvR/ClvR male, 30% is +/+ male, and 30% is +/+ female). Thus, initial release percentage also equals initial genotype frequency. Note that, for C and D, ClvR itself is assumed to have no fitness cost. Such costs would further increase the minimum release percentages required for drive to occur, as in A and B.
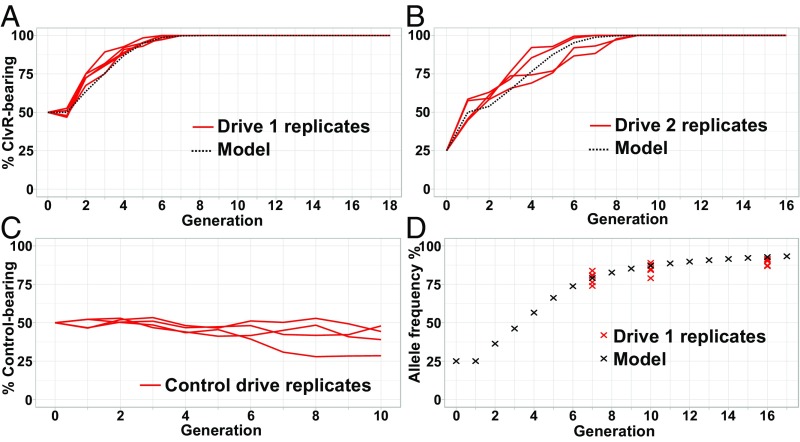
Fig. 5.
ClvR spreads to genotype fixation in Drosophila. The frequency of ClvR-bearing individuals (ClvR/+ and ClvR/ClvR) is indicated on the y axis and the generation number on the x axis. Drive replicates in red; predicted drive behavior in dotted black lines. (A) Drive 1: ♂ClvRtko/+ XX ♀w1118 as generation 0. (B) Drive 2: ♂ClvRtko/ClvRtko XX ♀ w1118 and ♂w1118 XX ♀w1118 at a 1:1 ratio as generation 0. (C) Control drive: ♂tkoA/+ XX ♀w1118 as generation 0. For the control drive, we used flies carrying construct tkoA (Methods) that had only the rescue and the td-tomato marker, but no Cas9 and gRNAs. (D) Allele frequency of ClvRtko in drive 1. Replicates coming from drive 1 in red. Model (black) is the predicted allele frequency inferred from modeling of the drive using parameters estimated from the data in Fig. 3, and assuming no fitness cost to carrying ClvR (see SI Appendix, Table S8, for counts).
Here, we focus on animals. When a heterozygous female mates with a wild-type male, female progeny survive because they inherit a wild-type copy of the essential gene from their father. Some males who inherit the X-linked LOF allele from their mother also survive because they inherit an autosomal copy of ClvR, while others die because they inherit the X-linked LOF allele and the wild-type non–ClvR-bearing autosomal homolog (Fig. 1C). If there is maternal carryover of Cas9/gRNA complexes, wild-type alleles of the essential gene inherited from the father can be converted to LOF alleles in the zygote. If this happens in a large fraction of nuclei in the zygote, all progeny not inheriting the ClvR-bearing chromosome, and thus lacking a functional copy of the essential gene, die (Fig. 1D). Together, these events create conditions in which ClvR-bearing parents transmit a potential fitness cost—a nonzero probability of inheriting no functional copies of the essential gene—to progeny. Non–ClvR-bearing homologous chromosomes are at risk for this cost, while ClvR-bearing chromosomes are not, thereby promoting a relative increase in frequency of the latter (Fig. 1 C and D).
Population Genetic Behavior of ClvR.
The behavior of such a ClvR element, located on an autosome and targeting a haplosufficient essential gene on the X chromosome (see Figs. 3 and 5 for related experiments), is illustrated in Fig. 2 for a conservative germline cleavage rate of 90% (actual rates, >99%; Fig. 3) and various release percentages and fitness costs, without (Fig. 2A) and with (Fig. 2B) 90% maternal carryover-dependent cleavage (actual rates, >99%; Fig. 3). ClvR is predicted to behave as a low-threshold gene drive mechanism (no deterministic threshold for an element with no fitness cost), spreading to transgene-bearing genotype fixation for a wide range of release percentages and fitness costs. However, in contrast to a HEG, which can spread quickly from low frequency (56), spread of ClvR is very frequency dependent: slow when introduced at low frequency, and fast when introduced at high frequency (Fig. 2 A and B). Maternal carryover-dependent cleavage is not essential for ClvR-dependent drive (Fig. 2A) but can speed the process and allow the drive element to tolerate larger fitness costs (Fig. 2B). Finally, while the behavior of many genes is described as haplosufficient, this designation often reflects the results of characterization under controlled laboratory conditions. Characterization of the same heterozygotes under other environmentally relevant conditions may uncover varying levels of haploinsufficiency (cf. ref. 57). Given that wild populations carrying gene drive elements will experience a variety of biotic and abiotic environmental conditions, it is important to understand how haploinsufficiency would affect ClvR-dependent drive. To explore this, we examined the behavior of a ClvR located on one autosome, targeting an unlinked locus on a different autosome, with a single functional version of the target gene resulting in some level of haploinsufficiency (Fig. 2C). We modeled a two locus autosomal scenario rather than that of an autosomal ClvR targeting the X since most essential genes are on autosomes, and to be able to capture the effects of haploinsufficiency in both sexes. Interestingly, ClvR is predicted to bring about population alteration under a wide variety of conditions if the essential gene targeted is haploinsufficient (Fig. 2C), or even haplolethal (Fig. 2D).
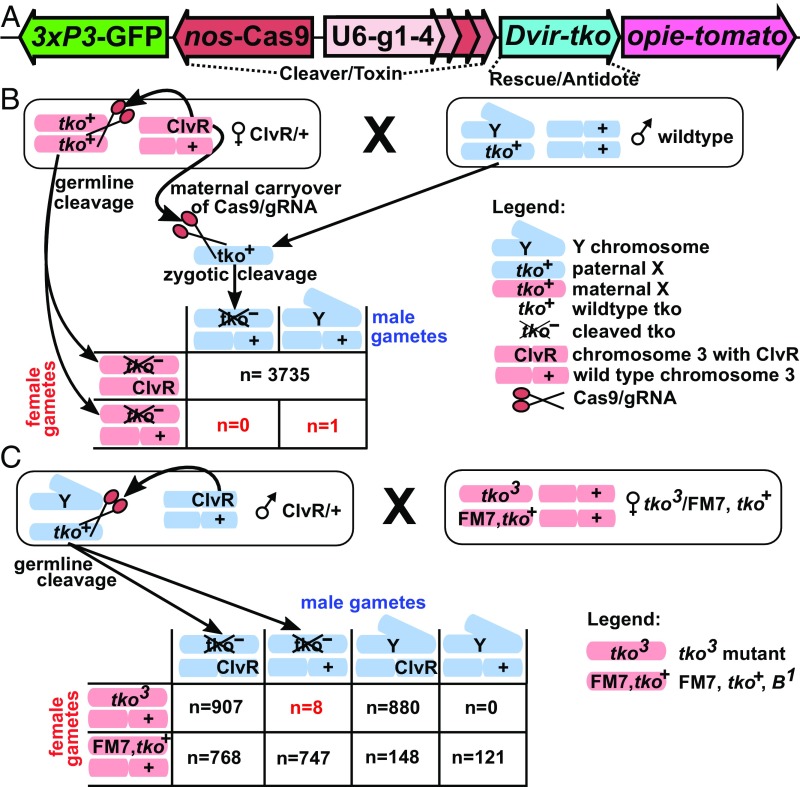
Fig. 3.
Components of ClvR and its behavior in females and males. (A) Component genes and their arrangement in ClvRtko. (B) The behavior of ClvRtko when present in a ClvRtko/+ adult female. Female progeny inherit an X from their mother (red) and one from their father (blue). Male progeny inherit an X from their mother. One non–ClvRtko -bearing male survived, while all other 3,735 male and female progeny inherited ClvRtko, for a cleavage rate of >99.9%. (C) The behavior of ClvRtko when present in a ClvRtko/+ male. When ClvRtko/+ males are crossed to tko3/FM7,B1 females (the FM7 balancer chromosome is wild type for tko), non-FM7,B1 female progeny carry tko3, a homozygous recessive lethal allele of tko) and an X chromosome from their father. In total, 907 of these carry ClvRtko, while only 8 (which may not represent independent events; SI Appendix, Fig. S5 and Table S3) do not, for a cleavage rate of >99%. Individuals carrying the FM7,B1 balancer, particularly males, are much less fit than others, and were not considered in the calculations.
Synthesis of ClvRtko in Drosophila melanogaster.
To create ClvR in Drosophila melanogaster, we first generated a construct carrying a recoded version of D. melanogaster’s X-linked tko locus, which encodes the conserved, essential, and haplosufficient mitochondrial ribosomal protein rps12 (58). To minimize homology of the rescue transgene with D. melanogaster tko, and thereby limit opportunities for recombination or gene conversion between the two (47, 48), we utilized the tko locus from a distantly related species, Drosophila virilis. We also introduced six additional silent coding sequence mutations to further reduce homology with the D. melanogaster gene (SI Appendix, Fig. S3). The tko rescue construct (tkoA) includes a dominant td-tomato marker, and an attP recombination site. It was introduced into the D. melanogaster genome on the third chromosome, at 68E, using Cas9 mediated HR, generating tkoA flies (SI Appendix, Fig. S4A). In a second step, transgenes expressing Cas9 and four gRNAs designed to recognize and cleave DNA within the D. melanogaster tko coding region, but not that of D. virilis tko, were integrated into the attP site in tkoA rescue construct-bearing flies (SI Appendix, Fig. S4 B and C). The gRNAs were each expressed under the control of a U6 polymerase III promoter (59). Cas9 was expressed under the control of nanos regulatory sequences, which drive expression in the male and female germline (60). Nanos-driven Cas9 also results in extensive maternal, but not paternal, carryover of active Cas9/gRNA complexes into the zygote (29, 61). The final construct is designated ClvRtko (Fig. 3A), and flies that carry it as ClvRtko flies.
Genetic Behavior of ClvRtko.
Matings between males that carry a LOF mutation for the X-linked eye pigmentation gene white (w1118), and that are heterozygous for ClvRtko on the third chromosome (w1118; ClvRtko/+), where + indicates a third chromosome that does not carry ClvRtko, and homozygous w1118; +/+ females resulted in high levels of progeny viability to adulthood (95.2 ± 2.0%), similar to those for the w1118 strain used for transformation (95.9 ± 2.0%). In addition, ∼50% (50.1 ± 3.0%) of the adult progeny carried ClvRtko, as expected for Mendelian segregation and high ClvRtko heterozygote fitness. Matings among homozygous ClvRtko flies also resulted in high levels of viability to adulthood (95.1 ± 1.7%), indicating that the presence of ClvRtko components (in the likely absence of functional D. melanogaster tko; see below) does not result in obvious fitness costs. In contrast, when heterozygous w1118; ClvRtko/+ females were mated with homozygous w1118; +/+ males, 53.6 ± 1.3% of progeny did not reach adulthood, and all surviving adults carried ClvRtko. On the basis of these results, we infer that the presence of ClvRtko in mothers results in a very high frequency (>99%) of mutational inactivation of the D. melanogaster tko locus in the adult female germline and in the zygote through maternal carryover-dependent cleavage of the paternal allele. In consequence, those who fail to inherit ClvRtko die, while those who inherit a single copy of ClvRtko thrive (SI Appendix, Table S1 A and B).
To obtain estimates of the rate of female adult germline- and maternal carryover-dependent cleavage and subsequent D. melanogaster tko inactivation, we repeated the cross between ClvRtko/+ females and wild-type males with larger numbers of individuals (see also SI Appendix, Table S5, for additional experiments of this type with genetically diverse strains). All but one of 3,736 progeny that survived to adulthood (cleavage rate of >99.9%) carried ClvRtko (Fig. 3B and SI Appendix, Table S2). To estimate male germline cleavage rates, we carried out a cross between ClvRtko/+ males and females that carried a lethal tko LOF allele [tko3, a frameshift mutation at amino acid 108 that introduces a premature stop codon (62)] in trans to the balancer chromosome FM7 (the balancer prevents meiotic recombination between X chromosomes), which is wild type for tko and carries a dominant mutation in the Bar gene, B1. Female progeny that inherit the maternal tko3 allele (identified by their failure to carry the dominant B1 marker), and that lack ClvRtko (and therefore lack the td-tomato and GFP markers), should die if D. melanogaster tko was inactivated in the parental male germline and survive if it was not. Eight females carrying the tko3 allele and lacking ClvRtko were recovered compared with 907 that carried tko3 and ClvRtko, for a minimum male germline cleavage rate of >99% (Fig. 3C and SI Appendix, Fig. S5 and Table S3). ClvRtko-dependent rescue of the tko3 mutant phenotype is indicated by the large numbers of tko3/Y; ClvRtko/+ progeny (880), compared with none for tko3/Y; +/+ (Fig. 3C).
X Chromosomes in Which a tko LOF Allele Was Not Created Following Exposure to ClvRtko Remain Sensitive to Cleavage by ClvRtko.
We sequenced the D. melanogaster tko locus from each of the nine X chromosomes above, in which a tko LOF allele was not created (escapers) following exposure to maternal or paternal ClvRtko. In the single escaper coming from a ClvRtko/+ mother, all four gRNA target sites were unaltered. For seven escapers coming from the ClvRtko/+ father, there was a common 3 bp in-frame deletion within the gRNA1 target site, and the remaining three target sites were unaltered. For escaper M3, a mixed sequencing signal, which may be indicative of nuclear mosaicism, was obtained. When each of the above escaper chromosomes was isolated in a male and the male crossed to ClvRtko/+ females, all surviving progeny inherited the ClvRtko td-tomato and GFP markers, showing that the D. melanogaster tko locus remained sensitive to cleavage (SI Appendix, Fig. S5 and Table S4).
ClvRtko Functions in Diverse Genetic Backgrounds.
To alter wild populations, a gene drive mechanism must be able to function in diverse genetic backgrounds. To begin to explore this topic with ClvR, we crossed ClvRtko/+ females to males from Global Diversity Lines (GDL) isolated from five different continents (63), and used in previous work investigating Cas9 function in the context of engineered HEGs (27). After each generation, we scored the frequency of ClvRtko flies, collected 30 virgins, and backcrossed them again to males from each of the GDL lines. Results are summarized in SI Appendix, Table S5. All offspring were ClvRtko-bearing for each of six generations (7,882 progeny scored). While these results do not preclude the existence of unlinked genetic variants and/or gRNA target polymorphisms in wild populations that would result in decreased rates of cleavage and LOF mutation creation, they show that the system is not specific to a common laboratory strain [SI Appendix, Table S6, shows all gRNA target site polymorphisms in strains from the 1000 fly genomes project (64)].
Molecular Nature of Mutations Created in D. melanogaster tko Created Following Exposure to ClvRtko.
To analyze the mutations in D. melanogaster tko created by ClvRtko we selected 2 ClvRtko-bearing male progeny from each of nine individual single crosses (18 total flies) between heterozygous ClvRtko females and w1118 males (from Fig. 3B). Sequencing results from the region of the D. melanogaster tko locus spanning the gRNA-binding sites are summarized in SI Appendix, Table S7A (alignments in SI Appendix, Fig. S6 A and B). The gRNA1 target site contained indels of varying size in all 18 individuals. The gRNA2 target site contained a likely preexisting polymorphism in four individuals (also observed in roughly half of the 1000 Fly Genome Project strains (64)), and a 2 bp deletion in 3. The gRNA3 target site was unaltered in all individuals, and the gRNA4 target contained indels in nine individuals. Somewhat surprisingly, larger deletions between target sites were not observed. This raises the possibility, suggested by others (65), that close juxtaposition of multiple target sites—in our case, four target sites within a 250-bp region of the tko ORF—limits Cas9’s ability to simultaneously interact with and/or cleave multiple nearby target sites as a consequence of Cas9-dependent DNA supercoiling.
One implication of such a model is that mutations should accumulate at additional target sites over time, as the target sites first cleaved by Cas9 are rendered nonfunctional for further Cas9 binding due to mutation within the gRNA target site. To explore this possibility, and the general question of whether all gRNA target sites can be cleaved, we sequenced the melanogaster tko locus from a homozygous ClvRtko stock that had been inbred for three generations (SI Appendix, Fig. S6 C and D, and Table S7B). Among the 12 analyzed males, all 12 had mutations at the gRNA1 target site. The gRNA2 target site was mutated in five, unaltered in one individual, and carried the suspected common polymorphism in the remaining six. The gRNA3 target site was mutated in 1 fly, and the gRNA4 target site was mutated in all 12 flies. Thus, cleavage events accumulate over time, and all sites can be cleaved, although cleavage efficiencies differ (from 100% for gRNA1 in generation 1 to 8% for gRNA3 after three generations).
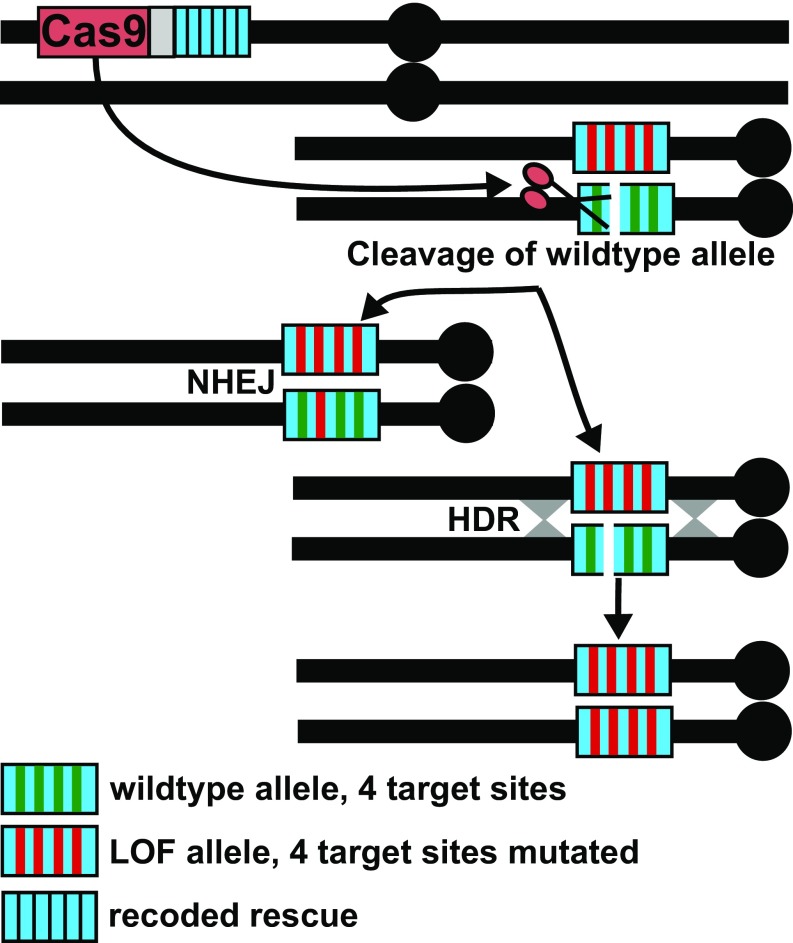
The mutations we observe presumably arise initially from error-prone repair by nonhomologous end-joining (NHEJ) or microhomology-mediated end-joining pathways (Fig. 4). However, we note that ClvR elements may also utilize HR and homing to create new LOF alleles when the ClvR-bearing individuals introduced into the wild population carry (as the above results indicate they will) uncleavable LOF indels in the targeted essential gene. For example, if ClvR-bearing individuals carrying LOF indels in the essential gene mate with wild-type, ClvR-bearing progeny will be heterozygous for chromosomes that carry the LOF indels and the wild-type version of the essential gene. In the germline of these individuals, the LOF indel-bearing chromosome (which is uncleavable) can serve as a template for HR-dependent repair of cleaved wild-type alleles, converting them to the LOF sequence (Fig. 4). Such behavior in cleavage heterozygotes was recently described in yeast (66). Further implications of homing-dependent alteration of the essential gene locus are discussed below.
Fig. 4.
LOF alleles can be created via cleavage followed by NHEJ, or via cleavage followed by HDR using an existing uncleavable LOF allele as a template for repair. The figure illustrates the germline of a female heterozygous for ClvR, and heterozygous for a LOF allele of the essential gene mutated at all four target sites, and a wild-type allele. Cleavage followed by error-prone repair (NHEJ) results in the creation of a new LOF allele mutated at one target site. Alternatively, cleavage can be followed by repair using the uncleavable LOF allele as a template, thereby resulting in conversion of the wild-type allele into a LOF allele in which all four target sites are mutated.
ClvRtko Spreads to Genotype Fixation in D. melanogaster.
Our combined results show that ClvRtko results in a very high frequency of germline and maternal carryover-dependent mutational inactivation of the D. melanogaster tko locus (>99% per generation); the lethality caused by this loss can be efficiently rescued using the D. virilis transgene; the high frequency of ClvRtko-dependent mutational inactivation of D. melanogaster tko and rescue by D. virilis tko is robust to genetic diversity; and cleaved but functional D. melanogaster tko alleles resistant to further cleavage, which could limit drive, were not observed. These observations predict that ClvRtko will spread to genotype fixation. To test this prediction, we initiated two drive experiments. In one experiment, w1118; ClvRtko/+ heterozygous males were mated with w1118; +/+ females, creating a progeny population used to seed the first generation in which ClvRtko was present in one-half of the individuals, at a total population allele frequency of 25%. In a second experiment, homozygous w1118; ClvRtko males and w1118; +/+ males were premated with equal numbers of w1118; +/+ females, which were then combined and used to seed the first generation (25% ClvR-bearing individuals), also resulting in an initial ClvRtko allele frequency of 25%. This level of introduction, although substantial, is not unreasonable as it is substantially lower than that used in earlier nontransgenic insect population suppression programs (67). As a control, we carried out similar drive experiments utilizing flies that carry the Rescue-only tko construct, tkoA, and that are wild type at the endogenous tko locus (w1118; tkoA). tkoA carries the td-tomato marker and the Rescue transgene, but lacks gRNAs and Cas9, and is thus expected to show Mendelian transmission. w1118; tkoA/+ males were mated with w1118; +/+ females (also wild type for tko), creating a progeny population used to seed the first generation in which tkoA was present in one-half of the individuals, at a total population allele frequency of 25%. For the first drive experiment, five replicate population cages were followed for 18 generations (drive 1, Fig. 5A). For the second drive experiment, four replicate populations were followed for 16 generations (drive 2, Fig. 5B). For the control, four tkoA populations were followed for 10 generations. In both ClvRtko drive experiments ClvRtko spread to genotype fixation between six and nine generations for all replicates. In contrast, the control transgene, tkoA, remained near its introduction frequency in all populations. As expected based on modeling, wild-type (+) alleles at the third chromosome locus into which ClvRtko was inserted were still present in the five drive 1 populations (Fig. 5D and SI Appendix, Table S8), but since wild-type alleles of D. melanogaster tko are eliminated by ClvRtko (SI Appendix, Fig. S6 and Table S7), these chromosomes are trapped in ClvRtko/+ heterozygotes.
Strategies for Maintaining ClvR Functionality over Time.
In any gene drive-based strategy for altering the makeup of a population, the cargo and drive mechanism are subject to separation, mutational inactivation, and loss of efficacy. Resilience, an ability to respond to these forces in ways that maintain and/or restore the ability to alter populations over time, is essential. Mutation of cargo genes or loss of effectiveness as a result of evolution of the host, or of other species such as pathogens on which they are meant to act, requires that strategies be available for removing an old element from the population and replacing it with a new one. This can be achieved using an approach analogous to that proposed for synthetic Medea selfish genetic elements (6, 68), in which a second-generation ClvR, ClvRn+1, is located at the same site as the first-generation element, ClvRn, with ClvRn+1 targeting essential genen+1, while also carrying the original rescuing copy of essential genen. Because progeny carrying ClvRn are sensitive to loss of essential genen+1, only those carrying ClvRn+1 survive, regardless of their status with respect to ClvRn (SI Appendix, Fig. S7). Opportunities for physical separation of Cargo from a functional Rescue can also be minimized, as with Medea (6), by interleaving Cargo and Rescue transgenes in various ways (SI Appendix, Figs. S8–S10).
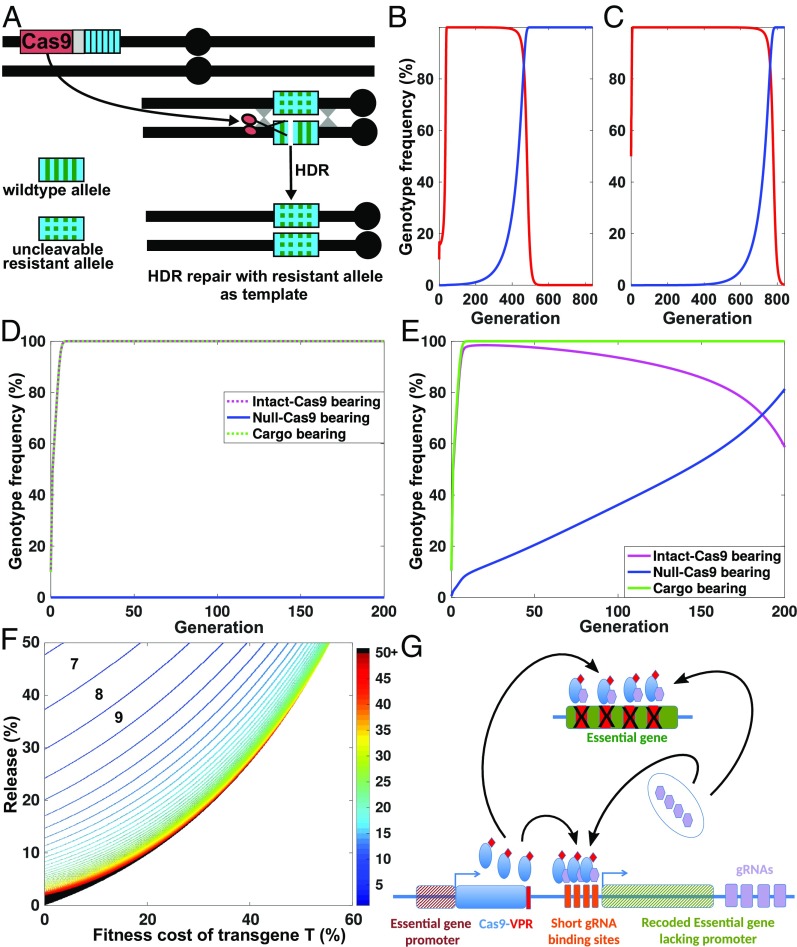
Cleavage is required for ClvR selfish behavior, and can fail as a result of mutation within target sites or Cas9/gRNAs. Mutations within the target sites that create uncleavable, but functional alleles of the target locus (resistant alleles), can lead to loss of ClvR from the population if its presence is associated with a fitness cost. Resistant alleles can arise from de novo mutations, from preexisting natural variation in the population, and as a result of error-prone NHEJ or microhomology-mediated end-joining pathways. Error-prone repair is likely to be the most important because the mutation rate per nucleotide/per generation is low, ∼10−8 to 10−9 (69), and high-frequency preexisting mutations that produce target site resistance to cleavage can be avoided through sequencing of the target population. In contrast, NHEJ-mediated creation of resistance alleles following cleavage can occur frequently [>10−3 per generation (27, 70)], although use of targets sites that cannot easily mutate to resistance and high fitness may be able to reduce this frequency dramatically (20). Modeling suggests that the probability of completely resistant alleles emerging with a multiplex of gRNAs is approximately equal to that of the probability of resistant alleles emerging at all gRNA target sites simultaneously, that is, pn, where p is the probability of a single site mutating to resistance and n is the number of gRNAs/target sites (71). Thus, even for a high rate of single target site mutation to resistance of 10−2 to 10−3, resistant alleles at all target sites might be predicted to arise only infrequently (∼10−8 to 10−12) with a four-gRNA ClvR. However, this calculation assumes no standing variation in the population at any of these sites, that all gRNAs work equally well, and that ectopic gene conversion between the Rescue transgene and the cleaved allele can be completely prevented by recoding.
The results reported herein, using laboratory and global diversity strains (0 resistant alleles out of more than 11,000 progeny scored; Fig. 3 and SI Appendix, Table S5), along with other recent work on HEGs (29, 61), provide experimental support for the idea that multiplexing of gRNAs can prevent the creation of cleavage-resistant, but functional alleles. Use of target sites that cannot easily mutate to a cleavage-resistant but high-fitness genotype have also been used toward a similar end (20). Targeting highly conserved housekeeping genes such as tko supports both strategies. Nonetheless, given that drive in very large populations has not yet been attempted, we briefly consider a “worst-case” scenario involving resistant alleles, ClvR, and a panmictic population, to gain some feeling for the consequences of resistant alleles on ClvR lifetime. We suppose that alleles that are completely resistant to four gRNAs, and with high fitness, arise at a high frequency of 10−6 per generation, that the presence of ClvR results in a significant fitness cost of 20% when homozygous (10% when heterozygous), and that ClvR is introduced at a low (10%) or a high (50%) frequency. Under these conditions, ClvR-bearing individuals constitute ≥99% of the population for 456 generations when introduced at a frequency of 10%, and 713 generations when introduced at a frequency of 50%. If homing of resistant alleles into cleaved wild-type alleles in heterozygotes carrying ClvR is now included (Fig. 6A), ClvR lifetime at high frequency (≥99% transgene-bearing) is modestly reduced to 409 generations for a 10% introduction frequency (Fig. 6B) and 707 generations for a 50% introduction frequency (Fig. 6C). The effect of homing is limited because it requires the presence of a specific genotype, a ClvR-bearing mother carrying both a wild-type and a resistant allele at the essential gene locus (Fig. 6A), and the speed at which ClvR elements transform wild-type alleles into LOF alleles works to limit the frequency of such individuals.
Fig. 6.
The consequences of target site/Cas9/gRNA inactivation for the spread of cargo, and ways of selecting against inactivation. (A) Illustration of how repair using HR and the resistant allele as a template can result in an increase in frequency of a resistant allele. (B) Genotype frequencies of ClvR (red line) and a resistant allele (blue) for an element that is introduced at a 10% genotype frequency (release of homozygous ClvR males), and that carries a 20% fitness cost, with 100% homing. (C) Same as in B, but for a 50% introduction frequency. (D) Behavior of ClvR for an element that is introduced at a 10% genotype frequency (release of homozygous ClvR males), and that carries a 10% fitness cost. No inactive versions of ClvR (dead Cas9) are present (0% null). (E) Same as in D, but with versions of ClvR that lack active Cas9 introduced, so as to make up 5% of the initial ClvR-bearing population. The fitness cost of dead Cas9-bearing ClvR elements is assumed to be half that (5%) of the fully functional element. (F) Heatmap showing number of generations needed for Cargo to reach transgene-bearing genotype fixation (>99%) for different release percentages and fitness costs, in which versions of ClvR that lack active Cas9 are introduced so as to constitute 5% of the ClvR-bearing individuals, for each release percentage. Fifty percent of the fitness cost associated with ClvR is assumed to be due to Cas9 activity, with the rest being due to Cargo. Thus, the wild-type, non-ClvR chromosome always has the highest fitness. Compare with Fig. 2B. (G) A hypothetical circuit that selects against mutation of Cas9/gRNAs to inactivity in which Cas9 activity is made essential for Rescue function.
With respect to mutational inactivation of Cas9 and gRNAs, ClvR-dependent drive of Cargo into a population is predicted to be remarkably insensitive to loss of these components when ClvR is introduced area-wide, even when inactive versions of Cas9/gRNAs are present at significant frequencies (5% of the ClvR-bearing individuals) in the initial population, and carriers of these mutant versions are more fit than those carrying intact Cas9 (Fig. 6 D–F). Drive is robust because so long as active ClvR elements are present, the population is rapidly driven toward Rescue- and thus Cargo-bearing genotype fixation by the ongoing loss of endogenous wild-type copies of the essential gene. Once all endogenous alleles of the essential gene are rendered nonfunctional, the population is locked into a Rescue—and thus Cargo-bearing—state regardless of whether Cas9 and gRNAs are still active. These points notwithstanding, we note that ClvR dynamics in the presence of resistant alleles at the target site or inactive Cas9 are likely to be more complicated in spatially structured populations that also include migration of wild types, a topic that remains to be explored. Strategies for further constraining the ability of Cas9/gRNAs to mutate to inactivity that involve forcing Cas9 and gRNAs to bring about transcription of the rescue as well as cleavage of the essential gene can also be envisioned (Fig. 6G). In one such strategy, a Cas9-VPR fusion protein is utilized. Cas9-VPR mediates cleavage at full length target sites. Cas9-VPR can also bind truncated gRNA target sites and drive transcription of a nearby gene, but is unable to cleave these sites (72). In this way, the same gRNAs and Cas9 are used for cleavage of the endogenous version of the essential gene and transcription of the Rescue transgene.
Conclusions.
Our findings demonstrate that the genetic composition of a population can be rapidly altered using the relatively simple toolkit of components that make up a ClvR gene drive/selfish genetic element: a site-specific DNA-modifying enzyme such as Cas9 and the gRNAs that guide it to specific targets, sequences sufficient to drive gene expression in the germline (which need not be germline-specific), an essential gene to act as target, and a recoded version of the essential gene resistant to sequence modification and able to rescue the LOF condition. Highly conserved housekeeping genes such as tko that participate in universal cellular processes required for cell survival or maintenance of basic cellular functions are good candidates for use in implementation ClvR in diverse species since they are essential in most if not all species (44–46). Importantly, modeling shows that drive and the alteration of populations to transgene-bearing genotype fixation can be achieved regardless of whether the essential gene being targeted is haplosufficient or haploinsufficient. This is likely to be important since haploinsufficiency may be more common than appreciated, and the fitness of individuals heterozygous for a LOF allele, under conditions present in the wild, is rarely known in advance. Finally, in the case where LOF alleles in the essential gene are created as a result of cleavage (as opposed to cleavage-independent base editing), ClvR does not require utilization of a specific repair pathway.
An important feature of ClvR is that the rate at which it spreads is frequency dependent (Fig. 2), very slow when introduced at low frequency, and fast when introduced at high frequency. In consequence, ClvR is likely to be most useful when it can be introduced area-wide, rather than from a point source within a larger area of interest. More detailed modeling that takes into account features such as density dependence, migration, and spatial structure is required to fully understand ClvR behavior. There are several other important unknowns. First, it is unclear what the costs and consequences are of long-term expression of DNA sequence-modifying enzymes such as Cas9, and if selection for alleles at other loci that result in decreased expression and/or activity may occur. A related unknown is the extent to which diversity in genome sequence in wild populations at the target site or elsewhere will thwart cleavage at the target locus. Our failure to identify cleavage-resistant, but functional tko alleles among >11,000 progeny from crosses of heterozygous ClvR-bearing females to wild-type males from a laboratory strain and GDL strains from five continents are promising in this regard, but the level of diversity tested likely pales beside that present in wild populations of some species of interest (73, 74). The problem of sequence diversity is also faced by other drive mechanisms designed to alter populations, such as synthetic Medea (6), some versions of underdominance (15, 16), and HEG-based homing (17, 19), which rely on the recognition of specific nucleotide sequences for their mechanism of action. Only further work in genetically diverse populations of species of interest, in facsimiles of wild environments, will suffice to determine whether synthetic selfish genetic elements able to thrive in the wild can be created.
Methods
Target Gene Selection and gRNA Design.
We selected the tko gene on the X chromosome as the target for the ClvR system. It encodes an essential mitochondrial ribosome protein and is recessive lethal and haplosufficient (58). We used the benchling software suite to design gRNAs targeting the exonic regions of the gene at four positions, selected based on on-target activity ranking (75). An additional criteria was that the gRNAs have a mutated PAM in the rescue construct to avoid any potential off-target cleavage therein (see below).
Cloning of ClvR Constructs and Fly Germline Transformation.
All plasmids used in this work were assembled with standard molecular cloning techniques and Gibson assembly (76). All restriction enzymes, enzymes for Gibson Assembly mastermix, and Q5 polymerase used in PCRs were from NEB; gel extraction kits and JM109 cells for cloning were from Zymo Research. The DNA extraction kit was from Qiagen (DNeasy). The gRNA cassette and Cas9 were based on pCFD3(4)-dU6:3gRNA and pnos-Cas9-nos, which were a gift from Simon Bullock, Division of Cell Biology, MRC Laboratory of Molecular Biology, Cambridge, United Kingdom (77) (Addgene; #49410 and #62208) and modified as described previously (61). Construct A (SI Appendix, Fig. S4A) was inserted into the fly germline via Cas9-mediated HR. Construct B (SI Appendix, Fig. S4B) was integrated into an attP landing site within flies carrying construct A using the phiC31 site-specific integration system. Detailed procedures can be found in SI Appendix, Supplementary Methods. Construct sequence fasta files can be found in Dataset S1.
Fly Crosses and Husbandry of ClvRtko Flies.
Fly husbandry and crosses were performed under standard conditions at 26 °C. Rainbow Transgenics carried out all of the fly injections. Containment and handling procedures for ClvRtko flies were as described previously (61), with G.O. and B.A.H. performing all fly handling. Details are in SI Appendix, Supplementary Methods.
Data Availability.
All data are available in the main text or SI Appendix. ClvRtko flies are available on request to labs that will meet or exceed containment guidelines outlined in ref. 61.
Supplementary Material
Acknowledgments
We thank Marlene Biller and Alexander Sampson for technical assistance, and Jackson Champer and Andrew G. Clark for providing the GDL Drosophila strains. Stocks obtained from the Bloomington Drosophila Stock Center (NIH Grant P40OD018537) were used in this study. This work was carried out with support to B.A.H. from the US Department of Agriculture (USDA) National Institute of Food and Agriculture (NIFA) Specialty Crop Initiative, under USDA NIFA Award 2012-51181-20086. G.O. was supported by a research fellowship from the Deutsche Forschungsgemeinschaft (OB428/1-1). T.I. was supported by NIH Training Grant 5T32GM007616-39.
Footnotes
Conflict of interest statement: The authors have filed patent applications on ClvR and related technologies (US Application No. 15/970,728).
This article is a PNAS Direct Submission.
See Commentary on page 5849.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1816928116/-/DCSupplemental.
References
- 1.Gould F, Huang Y, Legros M, Lloyd AL. A killer-rescue system for self-limiting gene drive of anti-pathogen constructs. Proc Biol Sci. 2008;275:2823–2829. doi: 10.1098/rspb.2008.0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noble C, et al. 2016. Daisy-chain gene drives for the alteration of local populations. bioRxiv:10.1101/057307. Preprint, posted June 7, 2016.
- 3.Burt A, Deredec A. Self-limiting population genetic control with sex-linked genome editors. Proc R Soc B. 2018;285:20180776. doi: 10.1098/rspb.2018.0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burt A, Trivers R. Genes in Conflict: The Biology of Selfish Genetic Elements. 1st Ed Belknap Press; Cambridge, MA: 2008. [Google Scholar]
- 5.Braig HR, Yan G. The spread of genetic constructs in natural insect populations. In: Letourneau DK, Burrows BE, editors. Genetically Engineered Organisms: Assessing Environmental and Human Health Effects. CRC Press; Boca Raton, FL: 2001. pp. 251–314. [Google Scholar]
- 6.Chen CH, et al. A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science. 2007;316:597–600. doi: 10.1126/science.1138595. [DOI] [PubMed] [Google Scholar]
- 7.Marshall JM, Hay BA. Inverse Medea as a novel gene drive system for local population replacement: A theoretical analysis. J Hered. 2011;102:336–341. doi: 10.1093/jhered/esr019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marshall JM, Pittman GW, Buchman AB, Hay BA. Semele: A killer-male, rescue-female system for suppression and replacement of insect disease vector populations. Genetics. 2011;187:535–551. doi: 10.1534/genetics.110.124479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marshall JM, Hay BA. General principles of single-construct chromosomal gene drive. Evolution. 2012;66:2150–2166. doi: 10.1111/j.1558-5646.2012.01582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gould F, Schliekelman P. Population genetics of autocidal control and strain replacement. Annu Rev Entomol. 2004;49:193–217. doi: 10.1146/annurev.ento.49.061802.123344. [DOI] [PubMed] [Google Scholar]
- 11.Davis S, Bax N, Grewe P. Engineered underdominance allows efficient and economical introgression of traits into pest populations. J Theor Biol. 2001;212:83–98. doi: 10.1006/jtbi.2001.2357. [DOI] [PubMed] [Google Scholar]
- 12.Altrock PM, Traulsen A, Reeves RG, Reed FA. Using underdominance to bi-stably transform local populations. J Theor Biol. 2010;267:62–75. doi: 10.1016/j.jtbi.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Altrock PM, Traulsen A, Reed FA. Stability properties of underdominance in finite subdivided populations. PLoS Comput Biol. 2011;7:e1002260. doi: 10.1371/journal.pcbi.1002260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akbari OS, et al. A synthetic gene drive system for local, reversible modification and suppression of insect populations. Curr Biol. 2013;23:671–677. doi: 10.1016/j.cub.2013.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gokhale CS, Reeves RG, Reed FA. Dynamics of a combined Medea-underdominant population transformation system. BMC Evol Biol. 2014;14:98. doi: 10.1186/1471-2148-14-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reeves RG, Bryk J, Altrock PM, Denton JA, Reed FA. First steps towards underdominant genetic transformation of insect populations. PLoS One. 2014;9:e97557. doi: 10.1371/journal.pone.0097557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burt A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc Biol Sci. 2003;270:921–928. doi: 10.1098/rspb.2002.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esvelt KM, Smidler AL, Catteruccia F, Church GM. Concerning RNA-guided gene drives for the alteration of wild populations. eLife. 2014;3:e03401. doi: 10.7554/eLife.03401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noble C, Olejarz J, Esvelt KM, Church GM, Nowak MA. Evolutionary dynamics of CRISPR gene drives. Sci Adv. 2017;3:e1601964. doi: 10.1126/sciadv.1601964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kyrou K, et al. A CRISPR-Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat Biotechnol. 2018;36:1062–1066. doi: 10.1038/nbt.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Windbichler N, et al. A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature. 2011;473:212–215. doi: 10.1038/nature09937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan YS, Huen DS, Glauert R, Whiteway E, Russell S. Optimising homing endonuclease gene drive performance in a semi-refractory species: The Drosophila melanogaster experience. PLoS One. 2013;8:e54130. doi: 10.1371/journal.pone.0054130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan YS, et al. The design and in vivo evaluation of engineered I-OnuI-based enzymes for HEG gene drive. PLoS One. 2013;8:e74254. doi: 10.1371/journal.pone.0074254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thyme SB, et al. Reprogramming homing endonuclease specificity through computational design and directed evolution. Nucleic Acids Res. 2014;42:2564–2576. doi: 10.1093/nar/gkt1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simoni A, et al. Development of synthetic selfish elements based on modular nucleases in Drosophila melanogaster. Nucleic Acids Res. 2014;42:7461–7472. doi: 10.1093/nar/gku387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gantz VM, Bier E. Genome editing. The mutagenic chain reaction: A method for converting heterozygous to homozygous mutations. Science. 2015;348:442–444. doi: 10.1126/science.aaa5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Champer J, et al. Novel CRISPR/Cas9 gene drive constructs reveal insights into mechanisms of resistance allele formation and drive efficiency in genetically diverse populations. PLoS Genet. 2017;13:e1006796. doi: 10.1371/journal.pgen.1006796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gantz VM, et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci USA. 2015;112:E6736–E6743. doi: 10.1073/pnas.1521077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Champer J, et al. Reducing resistance allele formation in CRISPR gene drive. Proc Natl Acad Sci USA. 2018;115:5522–5527. doi: 10.1073/pnas.1720354115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Champer J, et al. Molecular safeguarding of CRISPR gene drive experiments. eLife. 2019;8:e41439. doi: 10.7554/eLife.41439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan G, Braig HR. Genetically Engineered Organisms. CRC Press; Boca Raton, FL: 2001. The spread of genetic constructs in natural insect populations; pp. 251–314. [Google Scholar]
- 32.Beeman RW, Friesen KS, Denell RE. Maternal-effect selfish genes in flour beetles. Science. 1992;256:89–92. doi: 10.1126/science.1566060. [DOI] [PubMed] [Google Scholar]
- 33.Lorenzen MD, et al. The maternal-effect, selfish genetic element Medea is associated with a composite Tc1 transposon. Proc Natl Acad Sci USA. 2008;105:10085–10089. doi: 10.1073/pnas.0800444105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ben-David E, Burga A, Kruglyak L. A maternal-effect selfish genetic element in Caenorhabditis elegans. Science. 2017;356:1051–1055. doi: 10.1126/science.aan0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seidel HS, et al. A novel sperm-delivered toxin causes late-stage embryo lethality and transmission ratio distortion in C. elegans. PLoS Biol. 2011;9:e1001115. doi: 10.1371/journal.pbio.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nuckolls NL, et al. wtf genes are prolific dual poison-antidote meiotic drivers. eLife. 2017;6:e26033. doi: 10.7554/eLife.26033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu W, et al. A large gene family in fission yeast encodes spore killers that subvert Mendel’s law. eLife. 2017;6:e26057. doi: 10.7554/eLife.26057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akbari OS, et al. Novel synthetic Medea selfish genetic elements drive population replacement in Drosophila; a theoretical exploration of Medea-dependent population suppression. ACS Synth Biol. 2014;3:915–928. doi: 10.1021/sb300079h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buchman A, Marshall JM, Ostrovski D, Yang T, Akbari OS. Synthetically engineered Medea gene drive system in the worldwide crop pest Drosophila suzukii. Proc Natl Acad Sci USA. 2018;115:4725–4730. doi: 10.1073/pnas.1713139115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mruk I, Kobayashi I. To be or not to be: Regulation of restriction-modification systems and other toxin-antitoxin systems. Nucleic Acids Res. 2014;42:70–86. doi: 10.1093/nar/gkt711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harms A, Brodersen DE, Mitarai N, Gerdes K. Toxins, targets, and triggers: An overview of toxin-antitoxin biology. Mol Cell. 2018;70:768–784. doi: 10.1016/j.molcel.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Hernández-Arriaga AM, Chan WT, Espinosa M, Díaz-Orejas R. Conditional activation of toxin-antitoxin systems: Postsegregational killing and beyond. Microbiol Spectr. 2014;2 doi: 10.1128/microbiolspec.PLAS-0009-2013. [DOI] [PubMed] [Google Scholar]
- 43.Eid A, Alshareef S, Mahfouz MM. CRISPR base editors: Genome editing without double-stranded breaks. Biochem J. 2018;475:1955–1964. doi: 10.1042/BCJ20170793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kachroo AH, et al. Evolution. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science. 2015;348:921–925. doi: 10.1126/science.aaa0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kachroo AH, et al. Systematic bacterialization of yeast genes identifies a near-universally swappable pathway. eLife. 2017;6:e25093. doi: 10.7554/eLife.25093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamza A, et al. Complementation of yeast genes with human genes as an experimental platform for functional testing of human genetic variants. Genetics. 2015;201:1263–1274. doi: 10.1534/genetics.115.181099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gloor GB, Nassif NA, Johnson-Schlitz DM, Preston CR, Engels WR. Targeted gene replacement in Drosophila via P element-induced gap repair. Science. 1991;253:1110–1117. doi: 10.1126/science.1653452. [DOI] [PubMed] [Google Scholar]
- 48.Tham KC, Kanaar R, Lebbink JH. Mismatch repair and homeologous recombination. DNA Repair (Amst) 2016;38:75–83. doi: 10.1016/j.dnarep.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 49.Cook RK, et al. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol. 2012;13:R21. doi: 10.1186/gb-2012-13-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simmons MJ, Crow JF. Mutations affecting fitness in Drosophila populations. Annu Rev Genet. 1977;11:49–78. doi: 10.1146/annurev.ge.11.120177.000405. [DOI] [PubMed] [Google Scholar]
- 51.Lindsley DL, et al. Segmental aneuploidy and the genetic gross structure of the Drosophila genome. Genetics. 1972;71:157–184. doi: 10.1093/genetics/71.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hägg P, de Pohl JW, Abdulkarim F, Isaksson LA. A host/plasmid system that is not dependent on antibiotics and antibiotic resistance genes for stable plasmid maintenance in Escherichia coli. J Biotechnol. 2004;111:17–30. doi: 10.1016/j.jbiotec.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 53.Braun RE, Behringer RR, Peschon JJ, Brinster RL, Palmiter RD. Genetically haploid spermatids are phenotypically diploid. Nature. 1989;337:373–376. doi: 10.1038/337373a0. [DOI] [PubMed] [Google Scholar]
- 54.Hime GR, Brill JA, Fuller MT. Assembly of ring canals in the male germ line from structural components of the contractile ring. J Cell Sci. 1996;109:2779–2788. doi: 10.1242/jcs.109.12.2779. [DOI] [PubMed] [Google Scholar]
- 55.Niedenberger BA, et al. Dynamic cytoplasmic projections connect mammalian spermatogonia in vivo. Development. 2018;145:dev161323. doi: 10.1242/dev.161323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beaghton A, et al. Requirements for driving antipathogen effector genes into populations of disease vectors by homing. Genetics. 2017;205:1587–1596. doi: 10.1534/genetics.116.197632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohnuki S, Ohya Y. High-dimensional single-cell phenotyping reveals extensive haploinsufficiency. PLoS Biol. 2018;16:e2005130. doi: 10.1371/journal.pbio.2005130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Royden CS, Pirrotta V, Jan LY. The tko locus, site of a behavioral mutation in D. melanogaster, codes for a protein homologous to prokaryotic ribosomal protein S12. Cell. 1987;51:165–173. doi: 10.1016/0092-8674(87)90144-9. [DOI] [PubMed] [Google Scholar]
- 59.Wakiyama M, Matsumoto T, Yokoyama S. Drosophila U6 promoter-driven short hairpin RNAs effectively induce RNA interference in Schneider 2 cells. Biochem Biophys Res Commun. 2005;331:1163–1170. doi: 10.1016/j.bbrc.2005.03.240. [DOI] [PubMed] [Google Scholar]
- 60.Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8:243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- 61.Oberhofer G, Ivy T, Hay BA. Behavior of homing endonuclease gene drives targeting genes required for viability or female fertility with multiplexed guide RNAs. Proc Natl Acad Sci USA. 2018;115:E9343–E9352. doi: 10.1073/pnas.1805278115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toivonen JM, et al. Technical knockout, a Drosophila model of mitochondrial deafness. Genetics. 2001;159:241–254. doi: 10.1093/genetics/159.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grenier JK, et al. Global diversity lines—a five-continent reference panel of sequenced Drosophila melanogaster strains. G3 (Bethesda) 2015;5:593–603. doi: 10.1534/g3.114.015883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lack JB, Lange JD, Tang AD, Corbett-Detig RB, Pool JE. A Thousand Fly Genomes: An expanded Drosophila genome nexus. Mol Biol Evol. 2016;33:3308–3313. doi: 10.1093/molbev/msw195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Farasat I, Salis HM. A biophysical model of CRISPR/Cas9 activity for rational design of genome editing and gene regulation. PLoS Comput Biol. 2016;12:e1004724. doi: 10.1371/journal.pcbi.1004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gorter de Vries AR, et al. Allele-specific genome editing using CRISPR-Cas9 is associated with loss of heterozygosity in diploid yeast. Nucleic Acids Res. December 5, 2018 doi: 10.1093/nar/gky1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dyck VA, Hendrichs J, Robinson AS. Sterile Insect Technique: Principles and Practice in Area-Wide Integrated Pest Management. Springer; Dordrecht, The Netherlands: 2005. [Google Scholar]
- 68.Hay BA, et al. Engineering the genomes of wild insect populations: Challenges, and opportunities provided by synthetic Medea selfish genetic elements. J Insect Physiol. 2010;56:1402–1413. doi: 10.1016/j.jinsphys.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haag-Liautard C, et al. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature. 2007;445:82–85, and erratum (2008) 453:128. doi: 10.1038/nature05388. [DOI] [PubMed] [Google Scholar]
- 70.Hammond AM, et al. The creation and selection of mutations resistant to a gene drive over multiple generations in the malaria mosquito. PLoS Genet. 2017;13:e1007039. doi: 10.1371/journal.pgen.1007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marshall JM, Buchman A, Sánchez C HM, Akbari OS. Overcoming evolved resistance to population-suppressing homing-based gene drives. Sci Rep. 2017;7:3776. doi: 10.1038/s41598-017-02744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kiani S, et al. Cas9 gRNA engineering for genome editing, activation and repression. Nat Methods. 2015;12:1051–1054. doi: 10.1038/nmeth.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dickson LB, et al. Exon-enriched libraries reveal large genic differences between Aedes aegypti from Senegal, West Africa, and populations outside Africa. G3 (Bethesda) 2017;7:571–582. doi: 10.1534/g3.116.036053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Anopheles gambiae 1000 Genomes Consortium Genetic diversity of the African malaria vector Anopheles gambiae. Nature. 2017;552:96–100. doi: 10.1038/nature24995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 77.Port F, Chen H-M, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci USA. 2014;111:E2967–E2976. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or SI Appendix. ClvRtko flies are available on request to labs that will meet or exceed containment guidelines outlined in ref. 61.