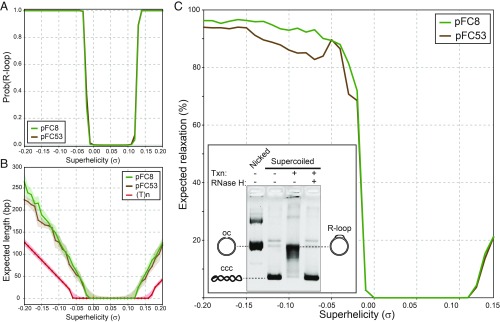
Fig. 3.
(A) The probability of R-loop formation is plotted as a function of superhelix density σ for the pFC8 (green) and pFC53 (brown) plasmids, as calculated by using R-looper. (B) Expected R-loop lengths are plotted for each plasmid as a function of σ. The shaded area around each curve delineates ±1 SD around the mean, shown as the central line. Since sufficient superhelicity will drive any sequence to form an R-loop, we included the expected length for the T homopolymer as a “negative” control. (C) This graph shows the percent relaxation of plasmid superhelicity expected from R-loop formation as a function of the initial template superhelix density σ for both pFC53 and pFC8. The expected relaxation is calculated as the fraction of superhelicity left after R-loop formation, (σ − σr)/σ. C, Inset shows the results of an in vitro transcription experiment in which negatively supercoiled pFC8 was transcribed to generate R-loops and treated with RNase H or not. R-loop formation was accompanied by a strong topological shift up to, or close to, the position of the relaxed, nicked substrate (open circle; oc). R-loop resolution by RNase H caused a full return of R-looped plasmids to the covalently closed circular (ccc) negatively supercoiled form.