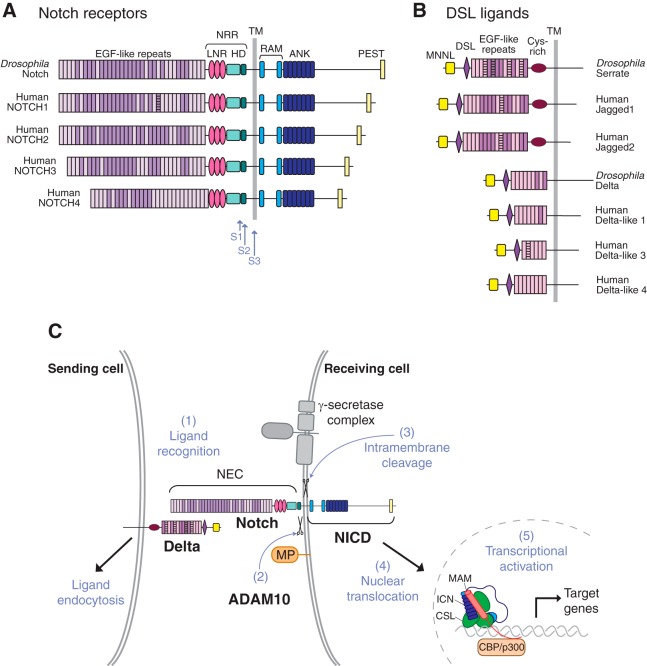
FIGURE 1.
Domain organization of Notch receptors and their membrane-anchored ligands and the role of proteolysis in Notch signaling. A and B: diagrammatic representation of the domain organization of the Drosophila Notch receptor and of the four human Notch receptors (A) and of the Drosophila and human Notch ligands (B). C: diagram depicting different steps involved in Notch signaling. A membrane-anchored Notch ligand, such as Delta, on the signal-sending cell, engages a membrane-anchored Notch receptor on the signal-receiving cell (1). This triggers endocytosis of the ligand, which provides the force to expose the Notch cleavage site to a disintegrin and metalloprotease 10 (ADAM10) for processing (2). This S2 processing step precedes the γ-secretase-dependent S3 processing event. This liberates the Notch intracellular domain (NICD) from its membrane anchor, allowing its translocation to the nucleus, where it enters into a transcriptional activation complex with CBF1/Su(H)/Lag-1 (CSL) and MAM, thereby activating transcription of Notch target genes. Please note that S1 processing by Furin in the secretory pathway is not shown in this diagram. ANK, ankyrin; HD, heterodimerization domain; LNR, Lin12/Notch repeat; NEC, Notch extracellular domain; NRR, negative regulatory region; PEST, proline-, glutamate-, serine-, and threonine-rich domain; RAM, RBP-Jκ-associated module; TM, transmembrane domain. [Adapted from Gordon et al. (83), with permission from J Cell Sci.]