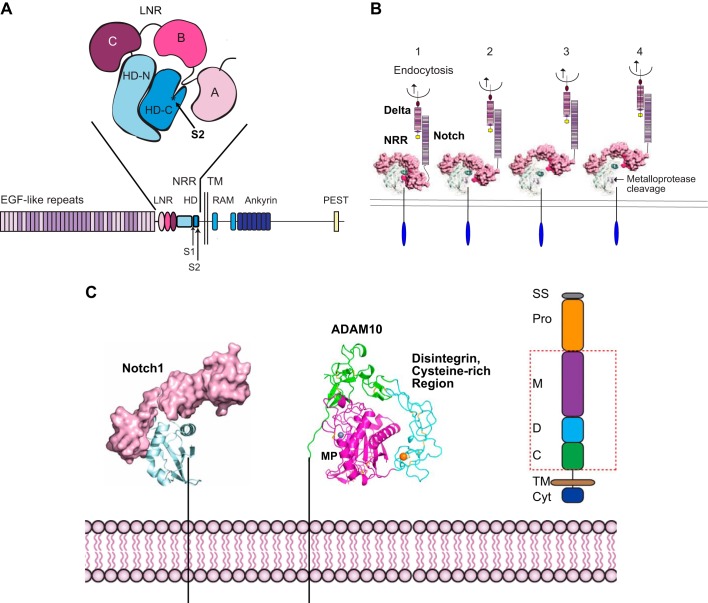
FIGURE 4.
Exposure of the Notch S2 cleavage site for a disintegrin and metalloprotease 10 (ADAM10) by endocytosis of the membrane-anchored Notch-ligand Delta. A: the negative regulatory region (NRR) of Notch, composed of three Lin12/Notch repeat domains (LNR; A, B, C) and a heterodimerization domain (HD-N, HD-C), which contains the buried S2 cleavage site for ADAM10 in HD-C. B: 1, endocytosis of a membrane-anchored Notch ligand such as Delta provides a force that is sufficient to unfold the NRR (85) (2–4), exposing the S2 ADAM cleavage site, indicated by an arrow in 4. C: once the cleavage site is uncovered, it can be processed by ADAM10 or ADAM17, generating a membrane-anchored stub that becomes a substrate for γ-secretase (see also FIGURE 1C). The structure of the extracellular domain of ADAM10, which is responsible for the processing of Notch under physiological conditions, is shown. The red box in the schematic of ADAM10 on the right indicates the domains shown in the structure of ADAM10. C, cysteine rich; Cyt, cytoplasmic tail; D, disintegrin; M, metalloproteinase; Pro, Pro domain; SS, signal sequence; TM, transmembrane. The colors in the schematic on the right correspond to those in the X-ray structure of ADAM10. The catalytic zinc ion is gray, and a bound calcium ion is shown in orange. Cysteine residues engaged in disulfide bonds are shown as sticks. [A is adapted from Gordon et al. (84), with permission from Blood; B and C from Gordon et al. (83), with permission from J Cell Sci; and C includes the structure of ADAM10, which was recently solved by Seegar et al. (215). We thank Stephen Blacklow for assistance in generating C.]