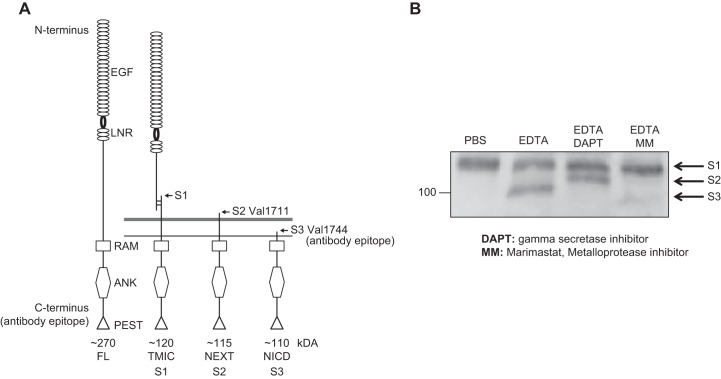
FIGURE 7.
Processing of Notch at the S1, S2, and S3 cleavage sites. A: diagrammatic representation of the uncleaved Notch1 precursor, as it is synthesized in the endoplasmic reticulum (left) (89). The S1 processing is a constitutive processing event carried out by furin-type pro-protein convertases in the Golgi apparatus. In physiological, canonical Notch signaling, S2 processing occurs in a ligand-dependent manner and depends on a disintegrin and metalloprotease 10 (ADAM10) (see also FIGURES 1 and 4). The generation of the juxtamembrane stub triggers further processing by γ-secretase, which releases the Notch intracellular domain (NICD) from its membrane anchor (see also FIGURE 1). B: Western blot analysis of nonphysiological EDTA-dependent activation of Notch processing, which depends on ADAM17 (20, 252), shows the S1, S2, and S3 cleavage products on a Western blot probed with an antibody against the cytoplasmic domain of Notch1 (Notch1 antibody, EP1238Y, Millipore). ANK, ankyrin; EGF, epidermal growth factor repeat; FL, full length; LNR, Lin12/Notch repeat; NEXT, Notch extracellular domain; RAM, RBP-Jκ-associated module; PEST, proline-, glutamate-, serine-, and threonine-rich domain; TMIC, transmembrane intracellular fragment. [A adapted from Groot et al. (89), with permission from J Invest Dermatol.]