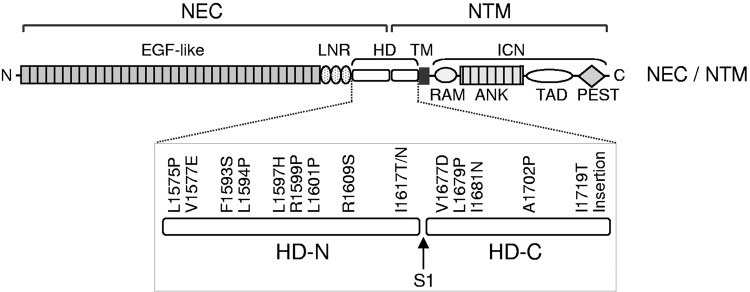
FIGURE 9.
A major class of oncogenic Notch mutations in the Notch negative regulatory region in T-cell acute lymphoblastic leukemia. Diagram of the location of mutations in Notch that cause cancer, many of which map to the negative regulatory region (NRR) domain [particularly in the heterodimerization domain (HD)]. This presumably leads to destabilization of the interacting domain units and to constitutive and inappropriate exposure of the Notch cleavage site, which can then be processed by both a disintegrin and metalloprotease 10 (ADAM10) and ADAM17 (233). Other mutations, such as those found in the proline-, glutamate-, serine-, and threonine-rich (PEST) sequence, affect turnover and stability of the Notch intracellular domain (NICD), thereby leading to increased and dysregulated Notch signaling. ANK, ankyrin; EGF, epidermal growth factor; ICN, intracellular Notch1; LNR, Lin12/Notch repeat; NEC, Notch extracellular domain; NTM, Notch1 transmembrane subunit; RAM, RBP-Jκ-associated module; TAD, transactivation domain; TM, transmembrane domain.[Adapted from Malecki et al. (160), with permission from Mol Cell Biol.]