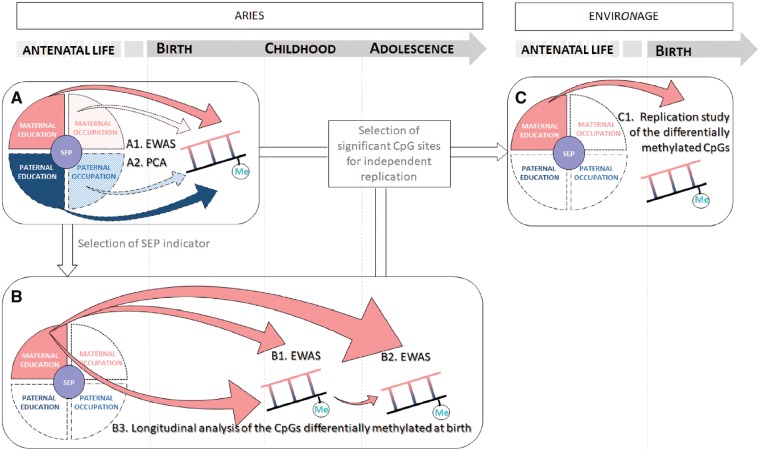
Figure 1.
Study workflow. The figure depicts the study workflow which is structured in three phases. First, the association between DNA methylation levels at birth and the four indicators of early life SEP was investigated performing EWAS (1 A1) and then regressing DNA methylation PCs against each indicators of SEP (1 A2). Second, based on significance from the PC analyses, a SEP indicator was selected. EWASs were performed for this selected indicator and DNA methylation status at childhood (1 B1) and at adolescence (1 B2), and methylation levels of the probes significant in cord blood were integrated over the three time points in a longitudinal model (1 B3). Finally, we adopted a targeted approach to seek independent validation of the CpG sites differentially methylated in relation to the selected SEP indicator both at birth and later in life, using neonatal biosamples from the ENVIRONAGE study (1 C1). EWAS, epigenome-wide association study; PCA, principal component analysis; SEP, socioeconomic position.