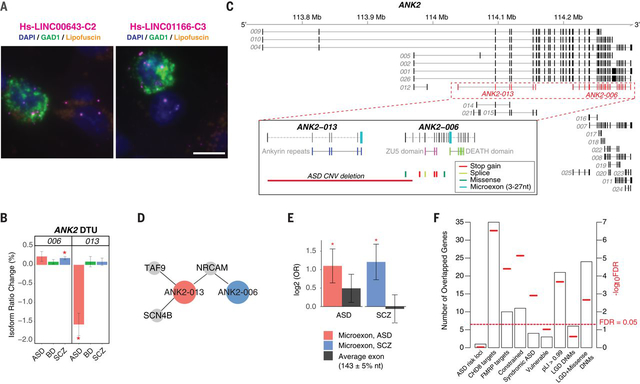
Figure 8. LncRNA annotation, ANK2 isoform switching & microexon enrichment.
A) FISH images demonstrate interneuron expression for two poorly annotated lincRNAs – LINC00643 and LINC01166 – in area 9 of adult human prefrontal cortex. Sections were labeled with GAD1 probe (green) to indicate GABAergic neurons and lncRNA (magenta) probes for LINC00643 (left) or for LINC01166 (right). All sections were counterstained with DAPI (blue) to reveal cell nuclei. Lipofuscin autofluorescence is visible in both the green and red channels and appears yellow/orange. Scale bar, 10 μm. FISH was repeated at least twice on independent samples (Table S9 (21)) with similar results (see also Fig S16). B) ANK2 isoforms ANK2–006 and ANK2–013 show significant DTU in SCZ and ASD, respectively (*FDR<0.05). C) Exon structure of ANK2 highlighting (dashed lines) the ANK2–006 and ANK2–013 isoforms. Inset, these isoforms have different protein domains and carry different microexons. ANK2–006 is hit by multiple ASD DNMs while ANK2–013 could be entirely eliminated by a de novo CNV deletion in ASD. D) Disease-specific coexpressed PPI network. Both ANK2–006 and ANK2–013 interact with NRCAM. The ASD-associated isoform ANK2–013 has two additional interacting partners, SCN4B and TAF9. E) As a class, switch isoforms are significantly enriched in microexon(s). In contrast, exons of average length are not enriched among switch isoforms. Y-axis displays odds ratio on log2 scale. P-values are calculated using logistic regression and corrected for multiple comparisons. F) Enrichment of 64 genes with switch isoforms in: ASD risk loci (81); CHD8 targets (96); FMRP targets (33); Mutationally constraint genes (97); Syndromic and highly ranked (1 and 2) genes from SFARI Gene database; Vulnerable ASD genes (98); Genes with probability of loss-of-function intolerance (pLI) > 0.99 as reported by the Exome Aggregation Consortium (99); Genes with likely-gene-disruption (LGD) or LGD plus missense de novo mutations (DNMs) found in patients with neurodevelopmental disorders (21).