Fig. 1. Phage selection and optimization of best Ni2+ cleavage sequence.
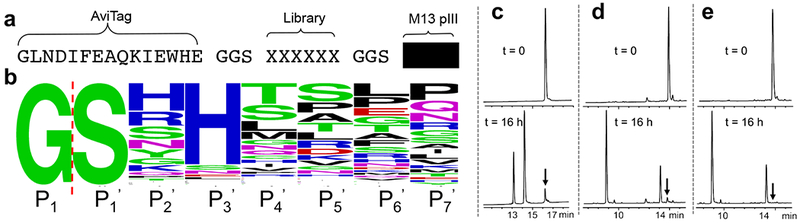
a. Substrate phage construct. b. Sequence frequency plot of Ni2+ cleavage selection, red dashed line indicates cleavage site between P1 and P1’ positions, P2’ to P7’ correspond to the randomized positions in Fig. 1a. Panels c-e. Evaluation of the cleavage of synthetic peptides (black arrow indicates uncleaved peptide). c. The most frequently observed phage selected peptide sequence YFLGGSHHTDLPGGSRRLFY; d. optimized peptide YFLPGSRHWG; e. best optimized peptide YFLPGSHHWG (the Arg for His substitution is based on the sequence logo in Fig. 1b). All peptides contain C-terminal carboxamides. The cleavage conditions were: peptide 0.2 mM, 0.1 M CHES, pH 8.2, 1 mM NiCl2, 22 °C, 16 hours. The mass of uncleaved and cleaved peptides were measured using MALDI-TOF shown in Supplementary Fig. 5.
