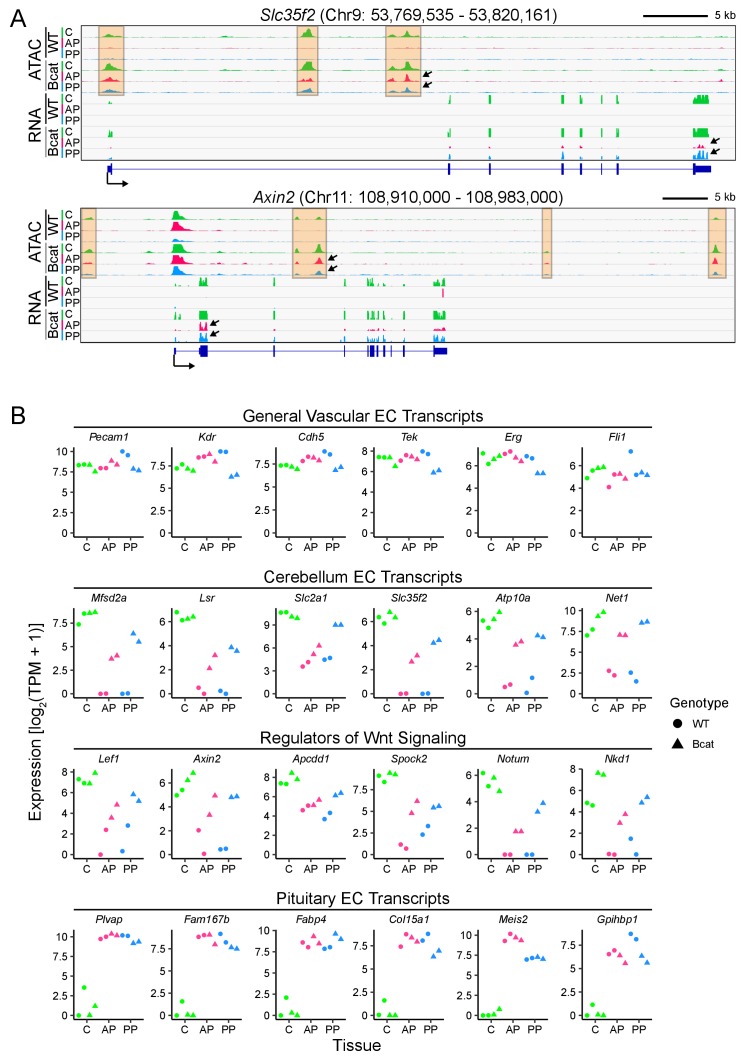
Figure 6. In pituitary ECs, stabilization of beta-catenin promotes a BBB-like state of the transcriptome and of chromatin accessibility.
(A) Genome browser images showing accessible chromatin (ATAC; top) and transcript abundances (RNA; bottom) for two BBB genes: Slc35f2, which codes for a putative nucleoside transporter, and Axin2, a Wnt target gene and a negative regulator of Wnt signaling. Histograms show aligned read counts. C, cerebellum; AP, anterior pituitary; PP, posterior pituitary; WT, wild-type; Bcat, beta-catenin stabilized (i.e. Ctnnb1 exon 3 deleted). Each track represents the average of two independent replicates. The orange regions highlight accessible chromatin that changes with beta-catenin stabilization. For each gene, all six of the ATAC-seq histograms are at the same vertical scale [Slc35f2 ATAC: 0–2000 reads; Axin2 ATAC: 0–3000 reads] and all six of the RNA-seq histograms are at the same vertical scale [Slc35f2 RNA: 0–5251 reads; Axin2 RNA: 0–3379 reads]. For both genes, some of the ATAC-seq and RNA-seq signals extend beyond the vertical range in the images. Bottom, intron-exon structure, with the arrow indicating the promoter and the direction of transcription. Arrows within the browser images point to examples of ATAC-seq or RNA-seq signals that are induced by beta-catenin stabilization. (B) Expression levels (log2 transformed TPM + 1) based on RNA-seq for the indicated categories of EC transcripts. Data for the two independent replicates are shown.
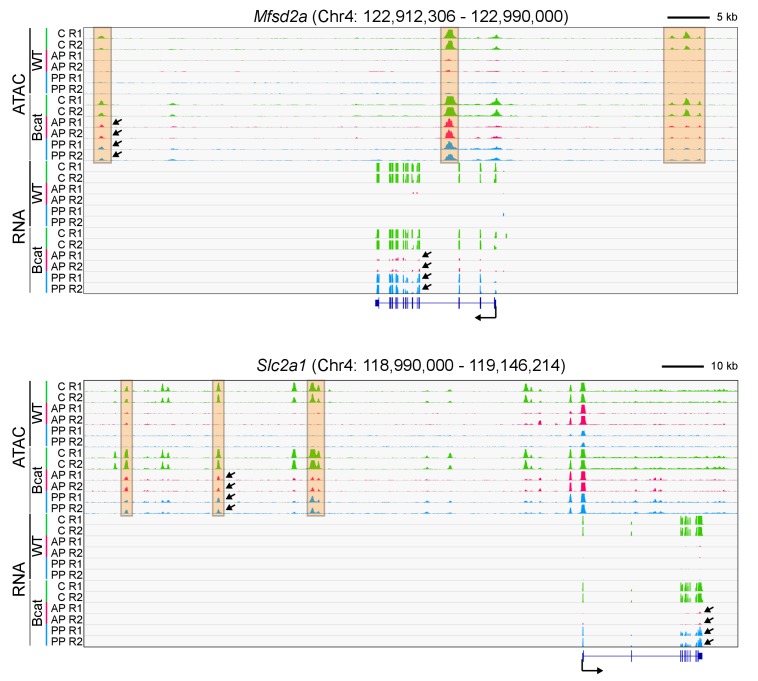
Figure 6—figure supplement 1. Genome browser images for two genomic loci that are responsive to beta-catenin stabilization in anterior and posterior pituitary ECs.
Figure 6—figure supplement 2. Heatmap showing the abundances of transcripts coding for tight junction proteins in WT vs.beta-catenin stabilized ECs in pituitary and cerebellum.
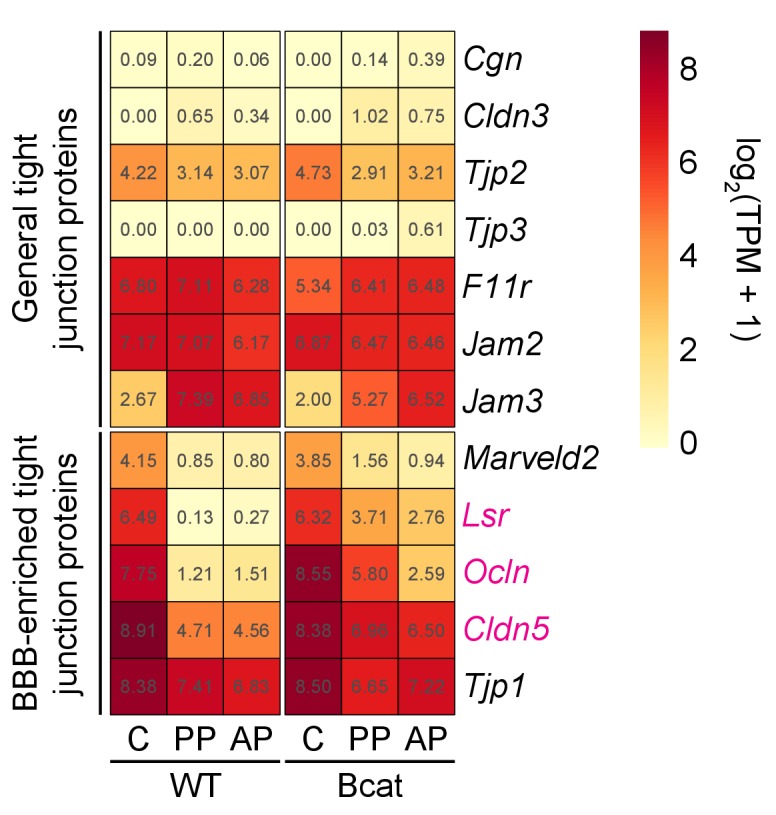
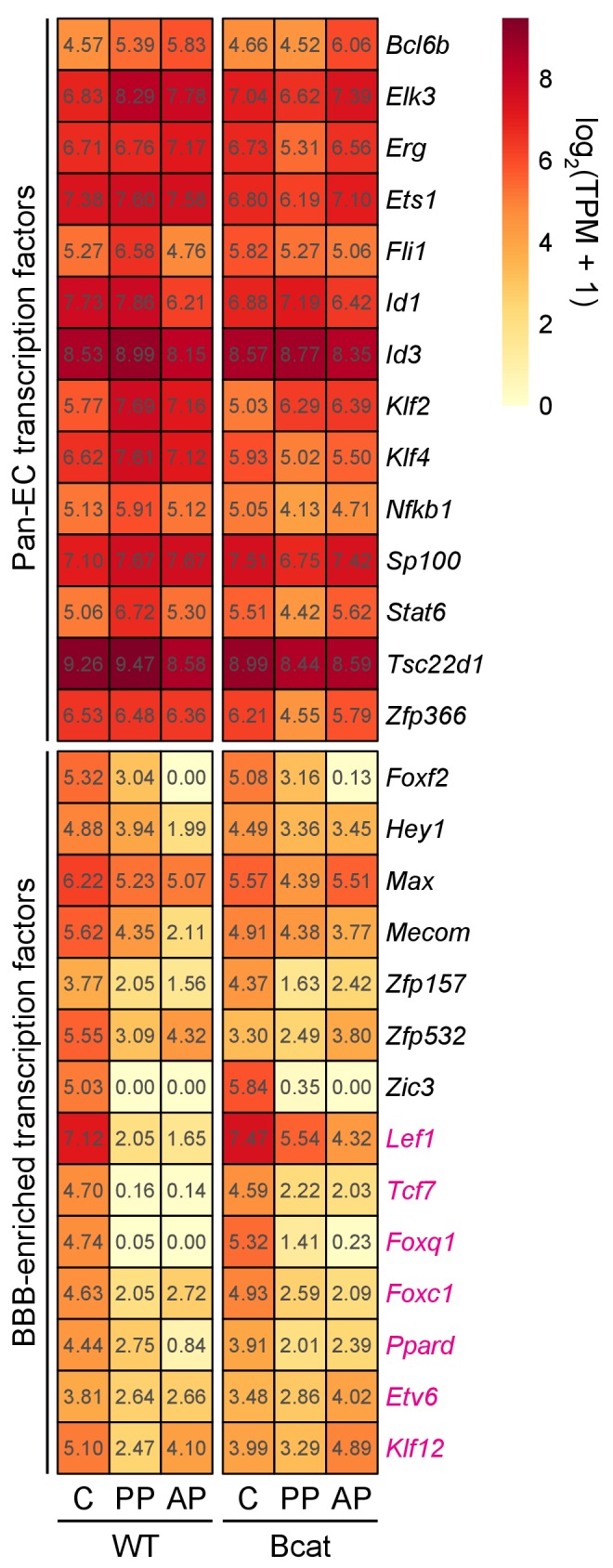
Figure 6—figure supplement 3. Heatmap showing the abundances of transcripts coding for TFs in WT vs. beta-catenin stabilized ECs in pituitary and cerebellum.