Figure 4: RNA-sequencing reveals a molecular HCM phenotype.
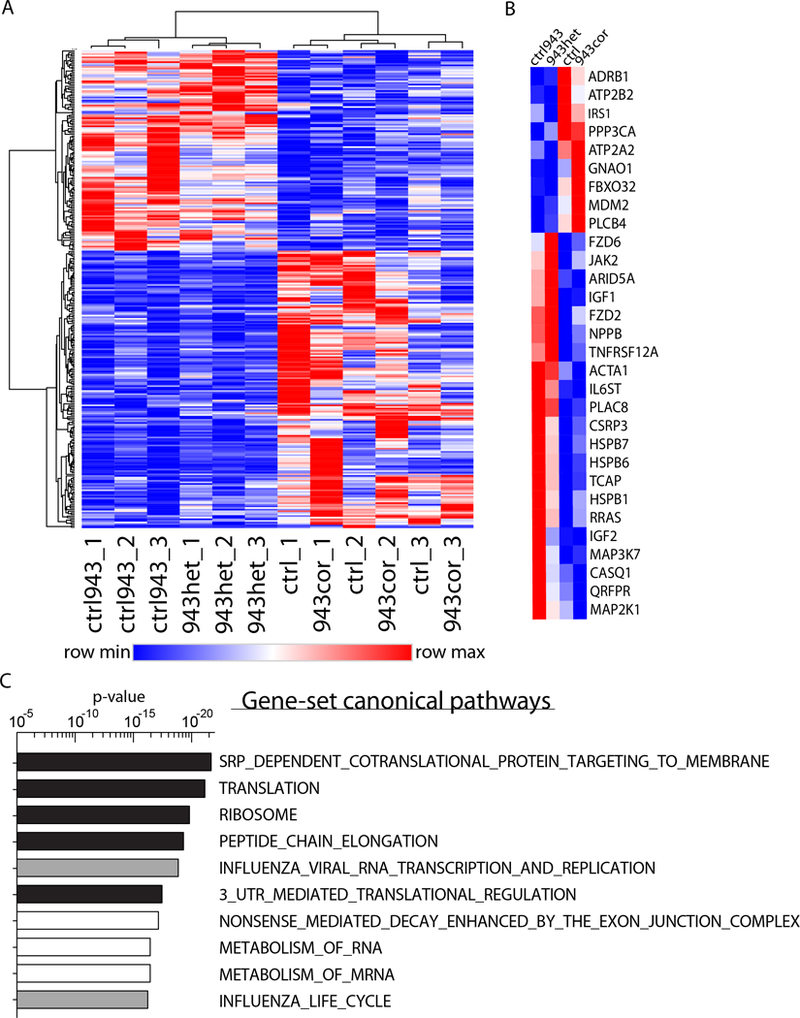
(A) Heatmap of the 298 significantly differentially regulated genes comparing HCM (943het, ctrl943) and healthy (943cor, ctrl) iPSC-CMs (fold-change ± 1.4; q-value <0.05; n=3 differentiation batches each iPSC line). 125 genes were upregulated and 173 genes were downregulated in HCM compared to healthy iPSC-CMs. (B) Within the top significant genes, a set of 30 genes were identified to be involved in cardiac hypertrophy, anabolic processes, and calcium signaling. (C) Top 10 canonical pathways activated in HCM iPSC-CMs identified by gene-set enrichment analysis (GSEA) of the genes. Pathways involved in translation are highlighted in black and pathways involved in RNA metabolism are highlighted in white.
