Figure 7.
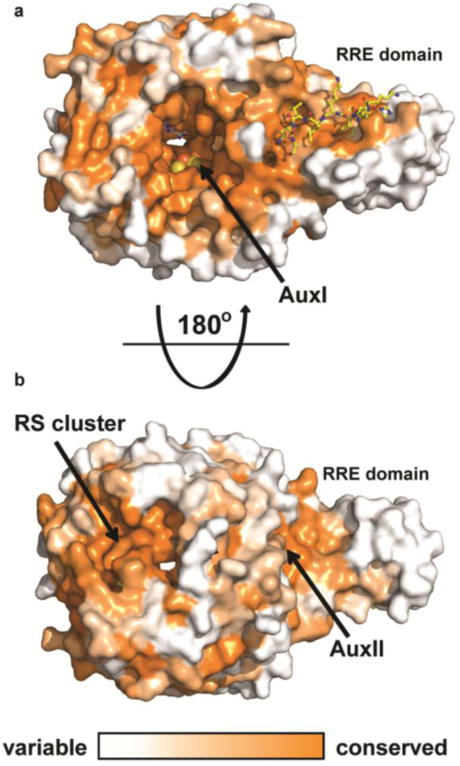
Conservation of CteB homologs. Surface map (ConSURF server) of sequence conservation based on 150 sequences with homology ranging from 35% to 90% identity. Conservation scores are based on Bayesian method. a) The highest sequence conservation can be found around the active site and peptide binding surface of the RRE domain. b) 180 ° rotation showing the bottom of CteB. A patch of highly conserved residues is found around the RS and Aux II clusters. These sites may have a role in the recognition of redox partners.
