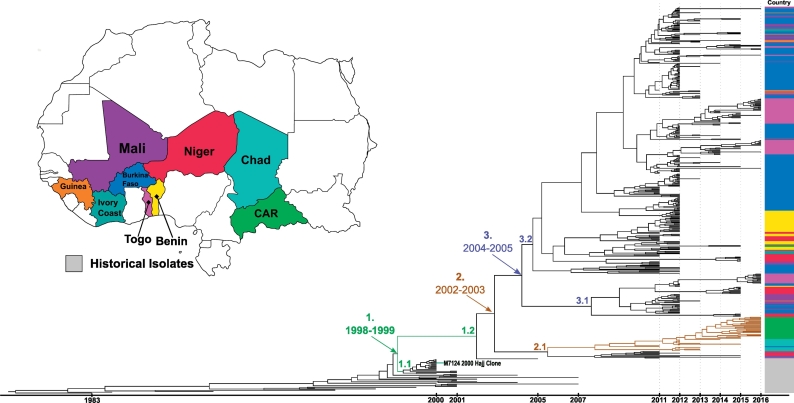
Fig. 2.
CC11 molecular clock timed phylogeny: A molecular clock based phylogeny was created for CC11 consisting of 434 study isolates and 40 historical (2007 or earlier) African isolates. The map represents the colour key for each country included in the phylogeny, with gray representing the historical isolates. The scale bar at the bottom indicates which year the isolates were collected from. Three clades of interest are labeled one to three, indicating the estimated date range for the origin of the common ancestor. The brown branches in sub-clade 2.1 represent a sub-lineage is primarily expanding in two Central African countries, Chad and Central African Republic. The 2000 Hajj outbreak clone (M7124) is also noted in the figure. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)