Table 1.
Optimization of the Reaction Conditionsa
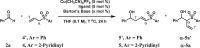 | |||||||
|---|---|---|---|---|---|---|---|
| Entry | Ar | Ligand | T (°C) | x | Total yieldb | γ/αb | ee of 5a′/5ac |
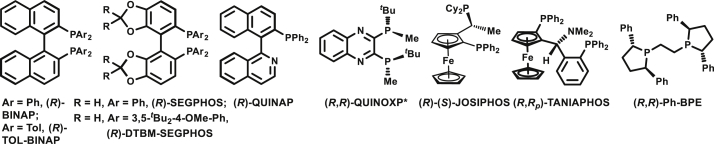
| 1 | Ph | (R)-BINAP | rt | 5 | 52% | 1/1.5 | 45% |
| 2 | Ph | (R)-TOL-BINAP | rt | 5 | 43% | 1/1 | 33% |
| 3 | Ph | (R)-SEGPHOS | rt | 5 | 42% | 1/1.5 | 8% |
| 4 | Ph | (R)-QUINAP | rt | 5 | 11% | 1/1 | 40% |
| 5 | Ph | (R,R)-QUINOXP* | rt | 5 | 48% | 1.5/1 | 8% |
| 6 | Ph | (R)-(S)-JOSIPHOS | rt | 5 | 51% | 1/1 | 58% |
| 7 | Ph | (R,Rp)-TANIAPHOS | rt | 5 | 47% | 1/2 | 51% |
| 8 | Ph | (R,R)-Ph-BPE | rt | 5 | 37% | 1/1 | 14% |
| 9 | Ph | (R)-DTBM-SEGPHOS | rt | 5 | 80% | >20/1 | 43% |
| 10 | 2-Py | (R)-DTBM-SEGPHOS | rt | 5 | 70% | >20/1 | 76% |
| 11 | 2-Py | (R)-DTBM-SEGPHOS | −40 | 5 | 76% | >20/1 | 97% |
| 12d | 2-Py | (R)-DTBM-SEGPHOS | −40 | 5 | 79% | >20/1 | 97% |
| 13d | 2-Py | (R)-DTBM-SEGPHOS | −40 | 30 | 92% | >20/1 | 97% |
| 14d,e | 2-Py | (R)-DTBM-SEGPHOS | −40 | 30 | 98% | >20/1 | 97% |
| 15d,e,f | 2-Py | (R)-DTBM-SEGPHOS | −40 | 30 | 82% | >20/1 | 97% |
HPLC, high-performance liquid chromatography; NMR, nuclear magnetic resonance
4/4′, 0.1 mmol; 2a, 0.2 mmol.
Determined by 1H NMR analysis of reaction crude mixture using mesitylene as an internal standard.
Determined by chiral-stationary-phase HPLC analysis.
4. 0.2 mmol; 2a. 0.1 mmol.
36 h.
Cu(CH3CN)4PF6 and ligand, 3 mol %, Barton's base = 2-tBu-1,1,3,3-tetramethylguanidine.