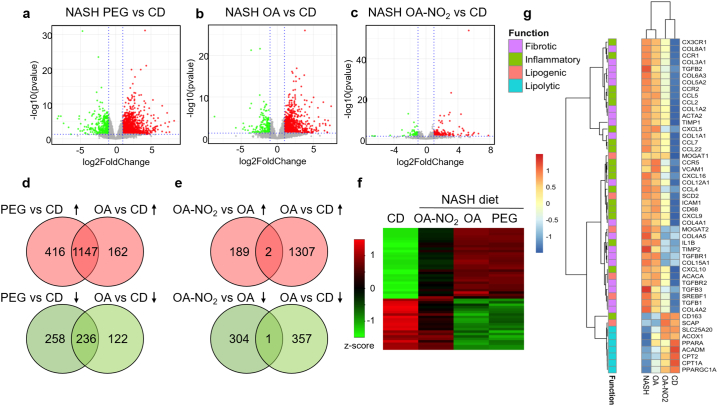
Fig. 4.
Global hepatic transcriptomic profile in response to OA-NO2 analyzed by RNA-sequencing. Volcano plots of differentially regulated genes (DEGs) in (a) NASH (PEG) vs. chow diet group (CD); (b) OA vs. CD and (c) OA-NO2 vs. CD. Principal component analysis (PCA) plot and additional volcano plot comparisons between groups are depicted in Supplementary Fig. 5a and b. X axis represents the log2FoldChange of each gene, and Y axis represents the -log10 transformation of the p values. Genes with p values <.05 and log2FoldChange larger than 1 were considered significant DEGs. Up-regulated DEGs are depicted in red, down-regulated DEGs in green, and the non-significant genes in grey. (d) Venn diagrams showing common DEGs in the NASH-diet (PEG) and OA vs. chow diet (CD). (e) Venn diagrams showing DEGs dissimilarity in the OA-NO2 group vs. OA. (f) Heatmap depicting the top 50 DEGs among all experimental groups as determined by log2FoldChage compared with CD group. Each row represents one gene, and each column represents one comparison to CD group. The log2FoldChange was row scaled and depicted by colors. (g) Heatmap of 50 NASH-related genes. The classification of the genes based on their function in inflammation, fibrosis, lipogenesis and lipolysis was labeled by different colors as shown at the left side of the heatmap. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)