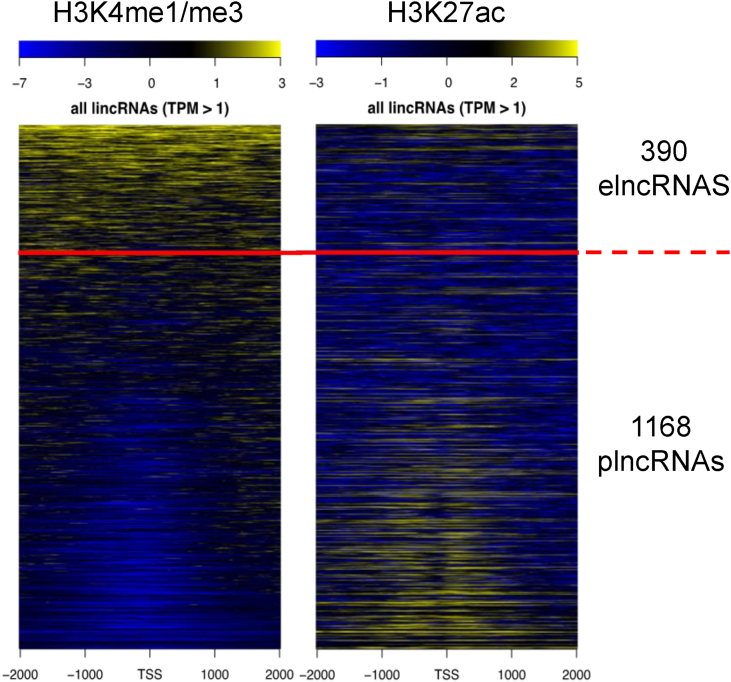
Fig. 4.
Histone ChIP-seq analysis of chondrocyte lincRNA. Histone ChIP-seq read alignment tracks for H3K4me3, H3K4me1 and H3K27ac were analysed from Roadmap cell type E049 (bone marrow-derived chondrocytes). Data presented are from the 1558 lincRNAs with a mean TPM > 1 in all hip and knee samples. Read coverage density is plotted around the TSS of lincRNAs (±2000 bp). H3K4me1 and H3K4me3 coverages were plotted as bam pairs using ngs.plot, with the same lincRNA order used to subsequently plot the H3K27ac read coverage. To define enhancer-associated (elncRNA) and promoter-associated (plncRNA) lincRNAs a threshold of 0.5 of H3K4me1/H3K4me3 was defined (red line).