Fig. 6.
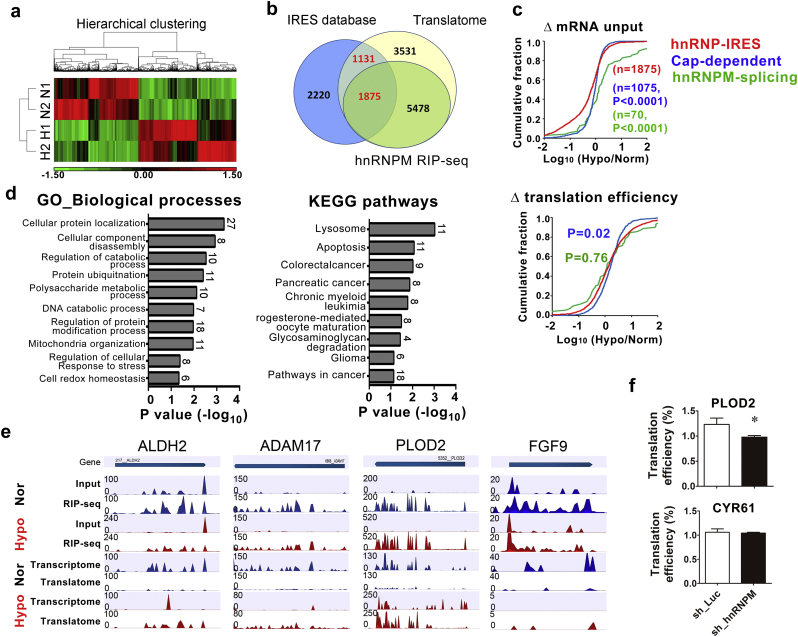
hnRMPM selectively controls IRES-mediated translation in response to hypoxia. (a) Hierarchical clustering of duplicated hnRNPM-IP-seq from cells exposed to normoxia (N1 and N2) or hypoxia (H1 and H2). Each column represents individual transcripts with significant (P < .05) up- (red) or down-expression (green). (b) Venn diagram shows overlapped targets from hnRNPMRIP-Seq (HEK293), translatome (HT-29), and predicted IRES-containing gene datasets. (c) Cumulative distribution log10-fold changes of mRNA input (upper) and translation efficiency (lower) among hnRNPM-IRES targets (red), hnRNPM-splicing targets (green), and cap-dependent translational targets (blue). P values were calculated using Mann-Whitney test (two-sided). (d) Gene ontology (biological processes, left) and KEGG pathways (right) analysis of 546 translation efficiency (TE) up-regulated targets. The number showing with each bar represents targets identified in each designated category. P value was presented as transformed –log10 value. (e) Genome browser view of the distribution of the RIP-seq and Ribo-Seq reads of represented genes. The upper panels show the hnRNPM-binding targets identified by RIP-seq (input, input RNA; RIP-Seq, hnRNPM immuno-precipitated RNAs), and the lower panels depict the Ribosome profiling (Ribo-Seq) reads of the represented genes (Transcriptome, input RNAs; Translatome, polyribome-bound RNAs). Normalized read densities of indicated dataset were shown for normoxia (blue) and hypoxia (red) (f) The quantitative measurement of mRNAs from hnRNPM-knockdown cytoplasmic extracts. Bar graphs represent the proportion of actively translated mRNA of PLOD2 (upper panel) and CYR61 (lower panel) from 3 independent experiments. ⁎P < .05 (Student's t-test). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)