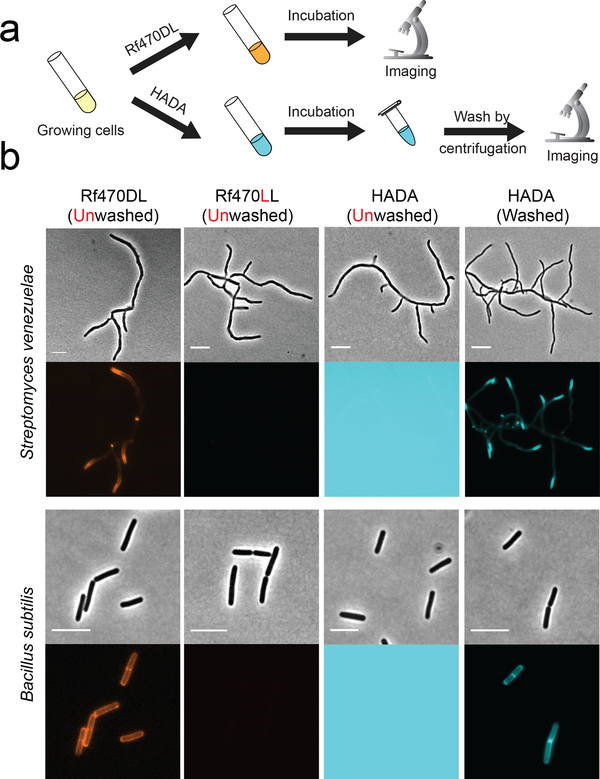
Figure 2. Unlike FDAAs, RfDAAs allow wash-free imaging of bacterial cell walls.
a) Scheme of labeling process. Bacterial cells were incubated with either RfDAAs or FDAA for PG labeling. The labeled cells were then collected and imaged with or without washing steps. b) Cell images shown in phase contrast (upper row) and fluorescence (bottom row) channels. Top panel: short-pulse labeling in S. venezuelae (1/3 doubling time); bottom panel: long-pulse labeling in B. subtilis (3 doubling times). PG labeling using RfDAAs (red) can be imaged without washing steps; whereas FDAA labeling (cyan) before washing steps results in saturated background fluorescence due to excess FDAAs in the field of view. Identical labeling, imaging and processing protocols were used for the comparison between D- and L- enantiomer labeling, as well as for unwashed and washed HADA labeling. Scale bar: 5 μm.