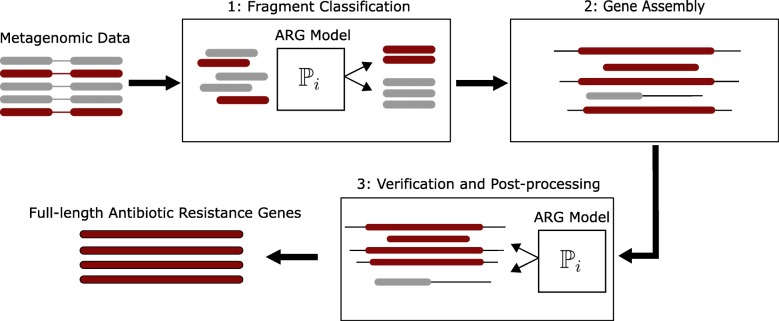
Fig. 1.
A schematic overview of fARGene. fARGene takes metagenomic paired-end data as input which then are subjected to an ARG model which classify the reads as coming from a resistance gene or not (panel 1). The paired-end sequences of the positively classified reads are extracted, quality controlled and then assembled into full-length genes (panel 2). The produced gene sequences are once again classified by the ARG model (panel 3). The output consists of nucleotide and amino acid sequences of the reconstructed ARGs. The method can also be applied directly to whole genomes and metagenomic contigs and then the classification, extraction, and assembly of reads are not performed