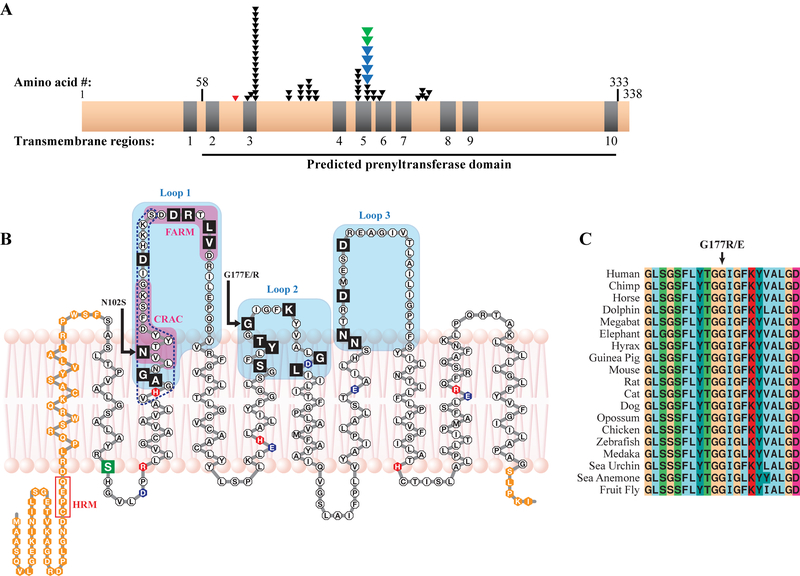
Figure 3.
SCD mutations and the UBIAD1 protein. A: Distribution of SCD family mutations on a linear protein model. Nine of ten predicted TM helices (grey) are in a predicted prenyltransferase domain (horizontal line, bottom) containing 19/19 residues altered in SCD families (one arrow per family), including p.G177E mutations in Swede-Finn (blue arrows) and Japanese and Turkish (green arrows) SCD families. A germline variant, p.S75F, is shown (red arrow). B: UBIAD1 in a lipid bilayer. SCD mutations (black boxes) occur in Loops 1–3 (shaded), including mutation hotspots asparagine 102 and glycine 177 (arrows). A predicted active site based on E.coli menA (dotted line, Weiss et al., 2008) and CRAC and FARM domains (Fredericks et al., 2011) are indicated. Acidic (blue), basic (red), a p.S75F SNP (green), and residues outside the predicted prenyltransferase domain (orange) are indicated. C: Complete conservation of codon p.G177 in 19 species.