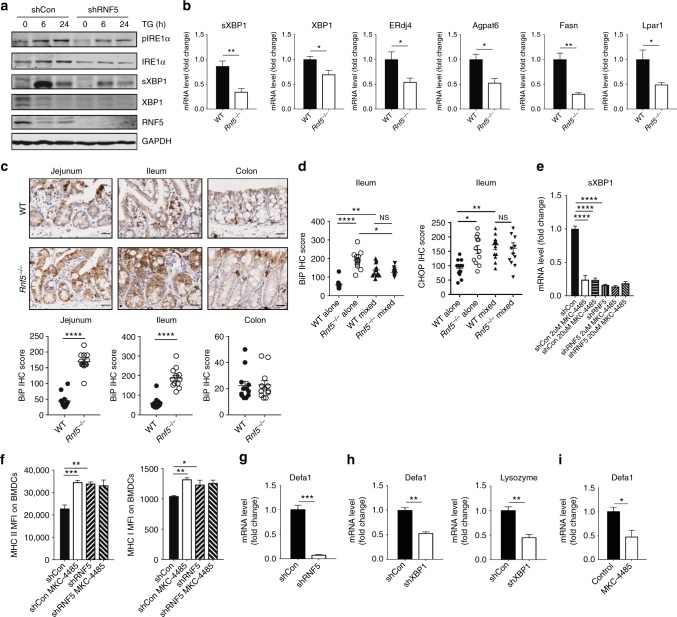
Fig. 6.
IEC of Rnf5−/− mice activate immune responses and changes AMPs expression by IRE1α/sXBP1 signaling. a Western blot analysis of the indicated proteins in lysates of MODE-K-shCon and MODE-K-shRNF5 cells treated with thapsigargin (TG, 1 μM) for indicated time. b qRT-PCR analysis of mRNA levels of sXBP1, XBP1 and target genes in small intestine-derived IECs from naive WT or Rnf5−/− mice (n = 4). c Representative micrographs of immunohistochemical (IHC) staining of BiP in the jejunum, ileum, and colon of YUMM1.5 tumor-bearing WT or Rnf5−/− mice (scale bar = 25 μm). Lower graphs show quantification of IHC staining (n = 12 fields per group). Staining was scored semi-quantitatively based on staining intensity (0, 1, 2, or 3) multiplied by the percentage of positively stained cells (0–100), to give a maximum IHC score of 300. d Quantification of BiP and CHOP IHC staining in ileum sections from WT and Rnf5−/− mice co-housed for 4 weeks prior to tumor inoculation (n = 12). e qRT-PCR analysis of sXBP1 mRNA levels in MODE-K-shCon and MODE-K-shRNF5 cells treated with the indicated concentration of MKC-4485 for 24 h (n = 3). f MFI of MHC class I and II on bone marrow-derived dendritic cells (BMDCs) incubated with media alone (no stimulation), conditioned media (CM) from shControl or shRNF5 MODE-K cells treated with MKC-4485 (n = 4). g qRT-PCR analysis of Defa1 mRNA levels in MODE-K-shCon and MODE-K-shRNF5 cells (n = 3). h qRT-PCR analysis of AMPs mRNA levels in IECs in MODE-K-shCon and MODE-K-shXBP1 cells (n = 3). i, qRT-PCR analysis of Defa1 mRNA levels in MODE-K cells treated with 2 μM MKC-4485 for 24 h (n = 3). Data are representative of three independent experiments (b, e–i) and two independent experiments (a, c, d) ≥3 mice per group. Graphs show the mean ± s.e.m. *P < 0.05, **P < 0.005, ***P < 0.001, ****P < 0.0001 by two-tailed t test or Mann–Whitney U test (b, c, g–i) or one-way ANOVA with Tukey’s correction for multiple comparisons (d–f)