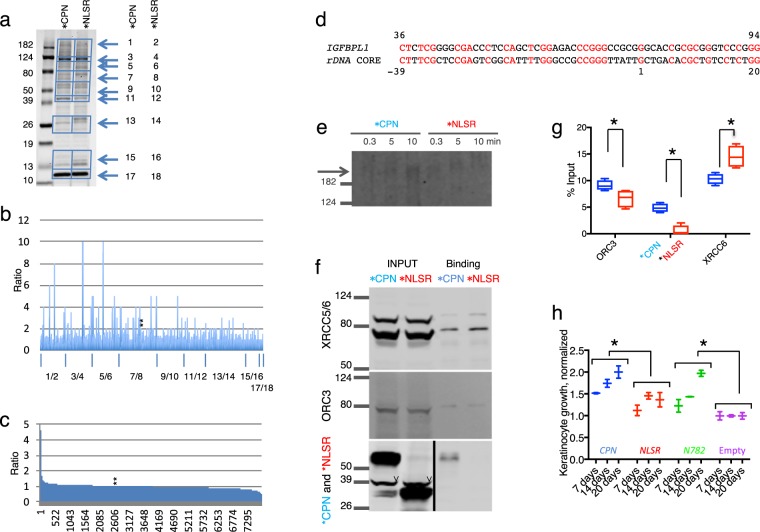
Figure 5.
EGFP-CPN enhances nuclear protein binding to the IGFBPL1-171 bait. (a) SDS-PAGE analysis of proteins that bound to the bait with EGFP-CPN-containing (*CPN) and EGFP-NLSR-containing (*NLSR) nuclear extracts. Slice identification number is shown on the right. (b) Mass spectrometry analysis of the bound proteins. Y-axis is ratio of bound-protein between the presence and absence of EGFP-CPN, and X-axis is the slice number. Two stars (**) show the ratio for ORC3 (2.0). (c) Mass spectrometry analysis of the EGFP-CPN-containing and EGFP-NLSR-containing nuclear extracts. Y-axis is the same as in (b), and X-axis is the protein identification number. Two stars (**) show the ratio for ORC3 (1.03). (d) Alignment of promoters of the IGFBPL1 and the rRNA genes. Nucleotides are numbered from the 5′ end of our clone for the IGFBPL, while TSS is shown by 1 in the rRNA promoter. (e) Time course of POLR1A binding (arrowed) to rDNA-260 promoter bait in the presence and absence of EGFP-CPN. 800 channel intensity, 6.5, was used to scan the image. Full-length blots are presented in Supplementary Fig. S4. (f) Binding of XRCC5/6, ORC3, and EGFP-CPN to the rDNA bait with the same extracts. (g) Quantification data of (f). 700 and 800 channel intensities were set to 4.5 and 6.5 for the scanning, respectively. * Shows a significant difference by t-test. p = 0.04 for ORC3, 0.01 for XRCC6, and <0.001 for *CPN vs *NLSR, n = 4. (h) Roles of CP on proliferation of keratinocytes. YF29 cultures were transduced with CPN (blue), NLSR (red), N782 (green), and the empty vector (violet) and cultivated for up to 20 days on 0.2 × 104/cm2 3T3-J2 (duplicates). * Shows a significant difference by ANOVA test (p = 0.0030 for CPN vs NLSR, and 0.0002 for N782 vs the control. Cell density of each measuring point was presented by fold over that of cultures transduced with the empty vector.