Fig. 4.
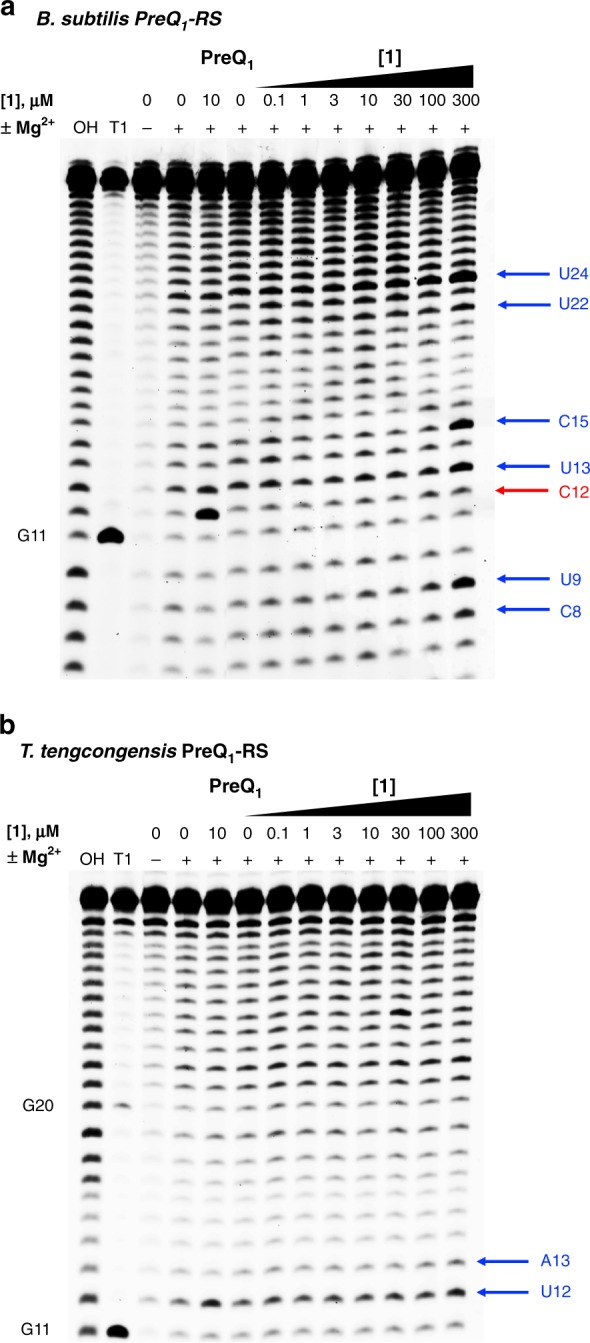
Ligand-induced conformational changes. In-line probing of a 5′-AlexaFluor 647-labeled BsPreQ1-RS RNA and b 5′-AlexaFluor 647-labeled TtPreQ1-RS RNA after treatment with 1 at increasing concentrations or a DMSO control in the absence (–) or presence (+) of 1 mm MgCl2. Treatment with PreQ1 at a concentration of 10 μm is used as a positive control. OH and T1 are a partial alkaline hydrolysis ladder and ribonuclease T1 digestion, respectively. Arrows designate nucleotide positions where the cleavage efficiency was significantly altered by compound treatment (blue) or preQ1 treatment (red)
