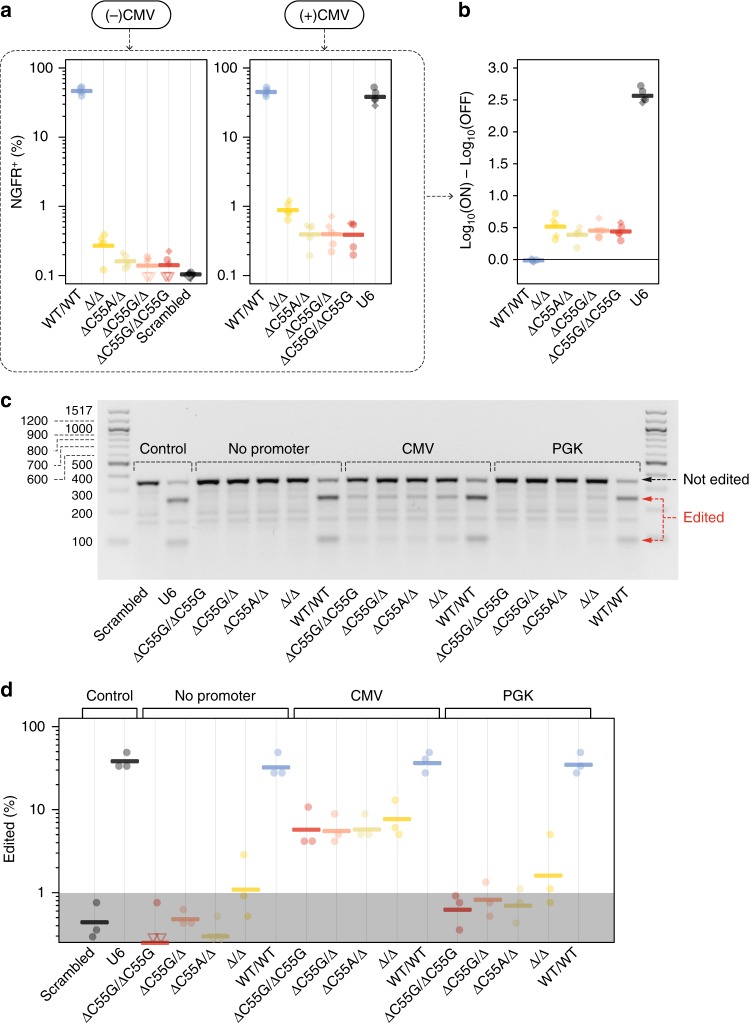
Fig. 4.
Engineered tRNAPro scaffolds enable Pol-II-specific endogenous gene targeting. a Percentage of NGFR+ cells within transfected cells for the OFF state ((−) CMV promoter) and the ON-state ((+) CMV promoter). Thick lines denote geometric mean values (n = 5 independent experiments; diamonds = day 4 with antibiotic selection; circles = day 3 no antibiotic selection; see Methods). Points at or below limit of detection are displayed as hollow triangles or diamonds for antibiotic selected experiments. Threshold of detection was set as containing ~0.1% of events in the scrambled gRNA control. b Log10(% NGFR+ cells) in the ON condition ((+) CMV promoter) compared to OFF condition ((−) CMV promoter). c Representative T7 endonuclease assay gel. tRNA scaffolds are shown beneath each plot with the 5′ variant/3′ variant indicated. A U6 promoter with a scrambled gRNA or with the targeting gRNA are included in as controls. Pol-II promoters for each set are noted above their respective lanes. Size in base pairs (bp) are shown next to the left side ladder. Black arrow = the location of uncut (not edited) amplicons; red arrows = digested (edited) amplicons. d Quantification of editing events from theT7 endonuclease gels. Thick lines represent geometric mean values (n = 3 independent experiments). Shaded area represents the 75% credible mass for the scrambled gRNA control (see also Supplementary Figure 7). Source data are provided as a Source Data file