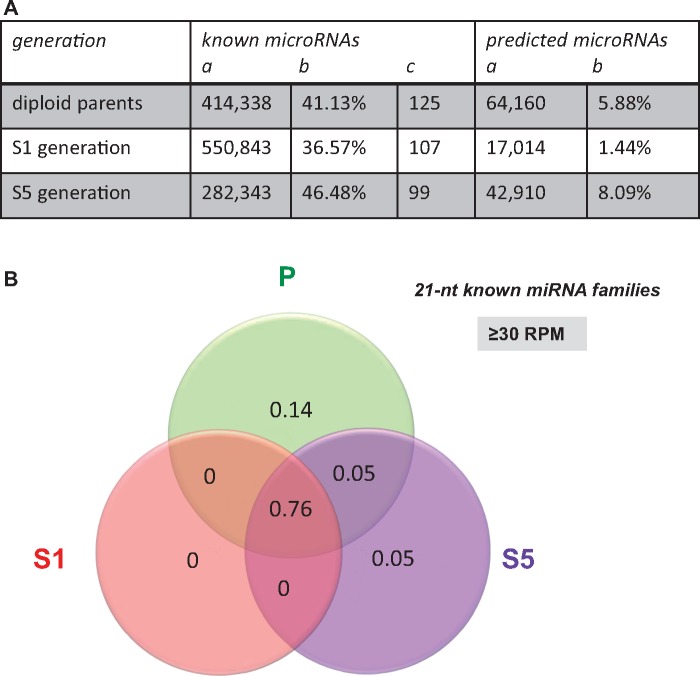
Fig. 3.
Characterization of the sRNAs annotated as miRNAs in the resynthesized Brassica napus allotetraploids surveyed at the two generations S1 and S5 in comparison with their diploid parents. (A) Summary table reporting the number of raw read counts (column a), the percentage of the 21-nt class (b) and the number of unique reads (c) corresponding to known or predicted miRNAs identified among the 21-nt reads. (B) Venn diagram showing the distribution of the 28 known miRNA families (according to miRBase v.19) that were present in the sequencing data with expression levels ≥30 RPM. Most of the miRNA families were found in the three different generations (76%); only five miRNA families were more abundant in the parental (14%) or the S5 generation (5%), or in both the parents and the S5 lines (5%). For this analysis, the sequencing data obtained from each library were pooled by generation: diploid parents “P” (pool of five libraries), S1 generation “S1” (3), and S5 generation “S5” (3). Distribution of known miRNAs appears thus as the opposite to the one depicted for the whole 21-nt class and reported in figure 2C.