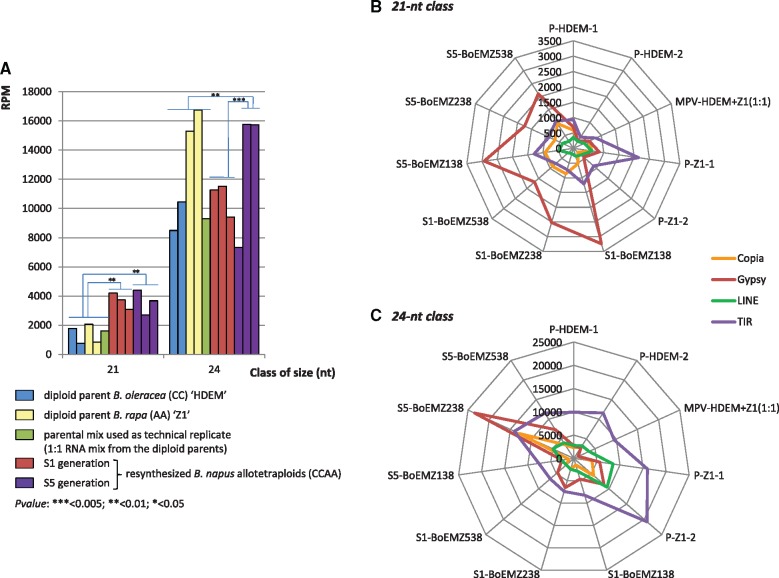
Fig. 5.
Differential expression of 21- and 24-nt siRNAs derived from TE sequences in the three resynthesized Brassica napus allotetraploids surveyed at the two generations S1 and S5 in comparison with their diploid progenitors. (A) The normalized abundances (in RPM) of siRNAs derived from LTR-retrotransposons were estimated for each of the 21- and 24-nt classes of sRNAs. We observed a significant increase of the 21-nt siRNAs in the S1 generation in comparison to the parental generation, which was maintained in the S5 generation. The 24-nt siRNAs displayed a significant increase only in the S5 generation in comparison to the parental and the S1 generations, which was particularly marked for the BoEMZ238 and BoEMZ538 neoallotetraploids. Spider graphs reporting comparisons of the expression levels (in RPM) of the 21-nt (B) and 24-nt siRNAs (C) derived from Copia and Gypsy LTR-retrotransposons across the different libraries to the expression levels of siRNAs derived from LINEs and TIR-transposons. They demonstrated the contrasting expression dynamics of siRNA populations according to the TEs the siRNAs originated from. Gypsy-derived 21-nt siRNAs were highly expressed in the S1 and S5 generations of the neoallotetraploids, whereas the other TE-siRNAs were mainly additively expressed. The Gypsy and Copia-derived 24-nt siRNAs were mostly expressed in the S5 generation, whereas the other TE-derived siRNAs were significantly underexpressed in the S1 generation. Annotation of TE-siRNAs was performed according to the TE classification published by Wicker et al. (2007).