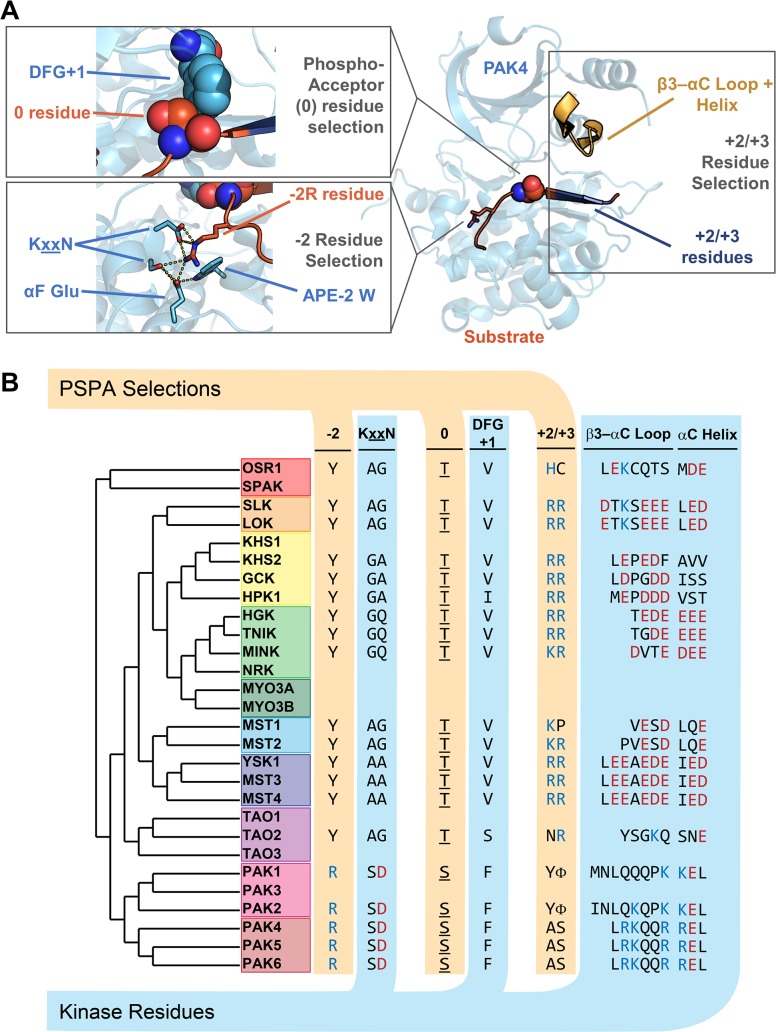
Fig 3. Overview of putative specificity-determining residues in the STE kinase family.
(A) Regions of the kinase proximal to the −2 (KxxN motif), 0 (DFG+1), and +2/+3 (β3–αC loop region) residues are highlighted on the x-ray crystal structure of PAK4 in complex with a peptide substrate (PDB: 2Q0N). (B) Dendrogram of the STE20 kinase family showing putative specificity-determining regions and corresponding residues selected by PSPA analysis. The alignment of the β3–αC loop region was made using data from available x-ray crystal structures and predictions from PSIPRED v3.3 [49, 50]. GCK, germinal center kinase; HGK, HPK/GCK-like kinase; HPK, Hematopoietic progenitor kinase 1; KHS, Kinase homologous to SPS1/STE20; LOK, Lymphocyte-oriented kinase; MINK, Misshapen-like kinase 1; MST, Mammalian sterile 20 kinase; MYO, myosin; NRK, NIK-related protein kinase; OSR1, Oxidative stress-responsive 1; PAK, p21-activated kinase; PDB, Protein Data Bank; PSPA, positional scanning peptide array; SLK, STE20-like kinase; SPAK, STE20/SPS1-related proline-alanine–rich kinase; TAO, thousand and one amino acid; TNIK, Traf2 and NCK-interacting protein kinase; YSK1, Yeast Sps1/Ste20-related Kinase 1