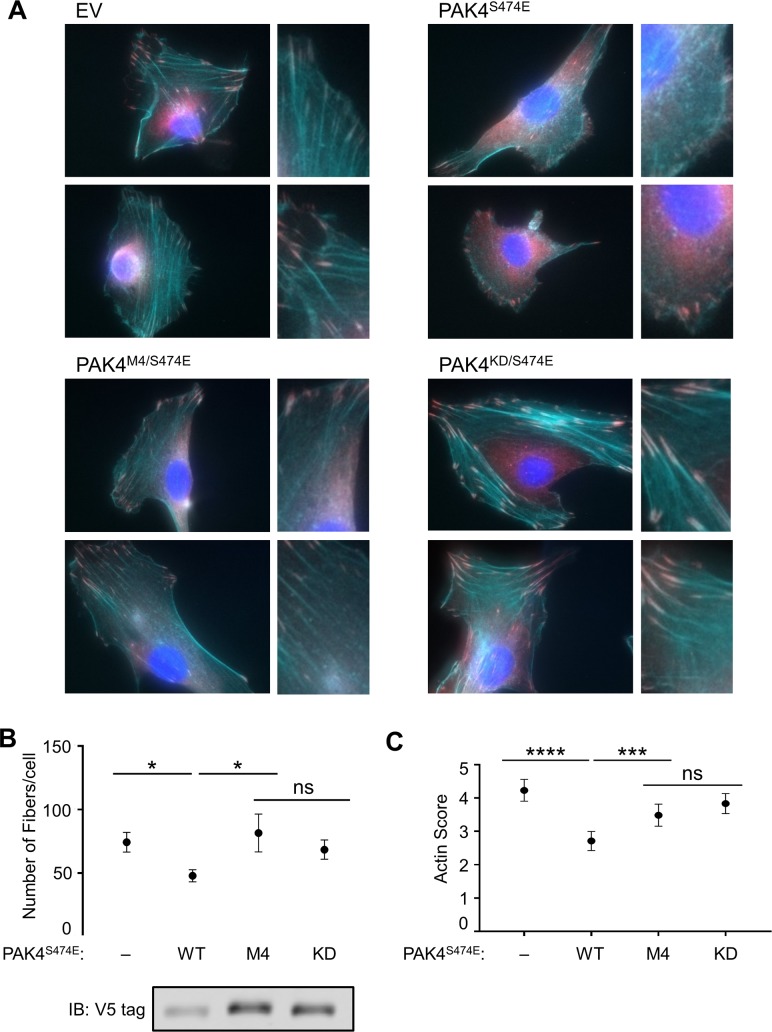
Fig 7. Phosphorylation site specificity is required for actin disassembly by PAK4.
(A) NIH-3T3 cells stably expressing the indicated PAK4 mutants were imaged following staining with phalloidin (cyan), α-vinculin (magenta), and DAPI (blue). (B) The number of actin fibers per cell were identified using SFEX software [54] to analyze blinded images. More than 30 cell images were analyzed for each condition. (C) Actin disassembly was scored manually on a 7-point scale in a blinded manner, with higher values indicating more actin fiber disassembly. At least 150 cells across three separate experiments were analyzed per condition. The scores of all four conditions were tested for differences using an ordinary, one-way ANOVA, which was significant (F = 16.99, p < 0.0001), followed by Fisher’s least significant difference post hoc test (***p < 0.001; ****p < 0.0001; ns, not significantly different at p = 0.05). Error bars indicate 95% CI. Cell lysates were immunoblotted to determine expression levels of V5-tagged proteins. Numerical data for (B) and (C) are provided in S2 Data. DAPI, 4′,6-diamidino-2-phenylindole; EV, empty vector; IB, immunoblot; KD, kinase-inactive mutant; PAK, p21-activated kinase; SFEX, stress fiber extractor; WT, wild type.