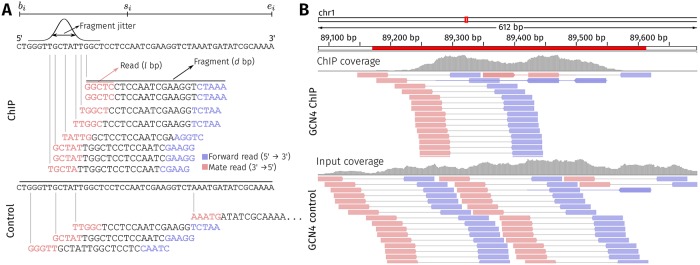
Fig 6. Example of simulating sequence reads using ChIPulate.
(A) Illustration of ChIPulate’s read generation method. The i-th genomic location is a genomic interval (bi, ei), with the “summit” of the interval located at si. The total read count (ri) and unique read count (ui) at the i-th interval in the ChIP and control samples are calculated as described in Fig 1. The starting positions of fragments in the ChIP sample are drawn from a Gaussian distribution with mean si − d/2 and standard deviation j, where d is the fragment length and j is the fragment jitter. In the control sample, fragment start positions are drawn from a Uniform(bi, ei) distribution. The Phred-33 quality value of every base is set to be 75, which corresponds to a base-calling error probability of 10−42. The read length is set to l bp, and both paired and single end reads can be simulated. (B) Paired-end reads simulated from genomic intervals containing an experimentally determined GCN4 binding site. The genomic intervals containing GCN4 binding sites were taken from an earlier publication [34]. Paired-end reads of 50 bp in length were simulated using ChIPulate, with the fragment jitter set at 50 bp and the fragment length set at 200 bp. The binding energy of the highest affinity GCN4 binding site in each interval was computed using the GCN4 binding energy matrix from the BEEML database. The remaining ChIPulate parameters were set at their default values.