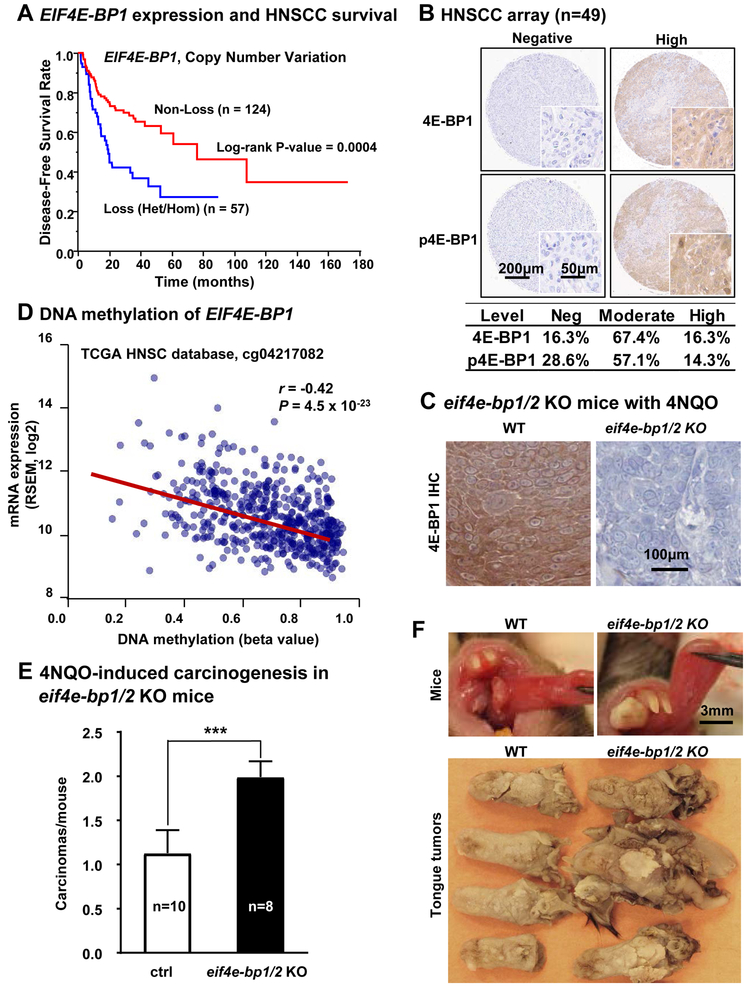
Figure 1: EIF4EBP1 is a candidate HNSCC tumor suppressor gene.
(A) The TCGA (The Cancer Genome Atlas) database was used to determine the relationship between EIF4EBP1 copy number variation (CNV) and disease-free survival (DFS). Data of CNV predicted by GISTIC algorism were available for 181 HNSCC patients in DFS (Log-rank test; p = 0.0004). Non-loss includes normal, copy gain and amplification. Loss includes heterozygous deletion and homozygous deletions. (B) Top, representative cores of HNSCC lesions stained with total 4E-BP1 (4E-BP1) and phospho-4E-BP1 (p4E-BP1, Thr37/46) using a HNSCC tissue microarray. Bottom, the intensity of staining was scored as previously described, (n=49) (6) and divided into negative, moderate and high expressed groups. (C) Representative immunohistochemical analysis of 4E-BP1 in WT and eif4ebp1/2 KO mice, respectively. (D) Significant negative correlation between DNA methylation and gene expression in the first intron of EIF4EBP1, suggestive of DNA methylation-mediated gene silencing (n=566, r = −0.42, p = 4.56 × 10−23). (E) 4NQO-induced carcinogenesis in eif4e-bp1/2 KO mice. Numbers of squamous cell carcinomas at the end of 4NQO-carcinogen treatment (mean ± SEM, n of WT mice = 10, n of 4e-bp1/2 KO mice= 8). (F) Top, representative pictures of live mice tongue in WT and eif4e-bp1/2 KO mice on week 26 of 4NQO treatment. Bottom, representative pictures of tongue lesions in WT and eif4e-bp1/2 KO mice on week 26 (time point when mice were sacrificed) of 4NQO treatment. These tumors in eif4e-bp1/2 KO mice were larger than those in wild type C57Bl/6 mice.