FIG 5.
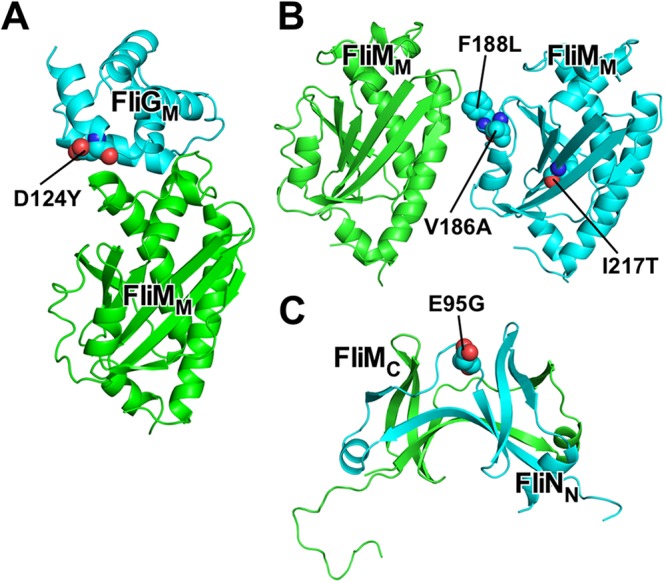
Locations of suppressor mutations of a FliF-FliG deletion fusion. (A) Homology model of Salmonella FliGM-FliMM complex and location of the FliG(D124Y) suppressor mutation. A homology model was built based on the crystal structure of Thermotoga maritima FliGM-FliMM complex (PDB code 3SOH). Cα ribbon drawing of FliGM (cyan) and FliMM (green). (B) Model of FliM subunit arrangement in the C ring and locations of the FliM(V186A), FliM(F188L), and FliM(I217T) suppressor mutations. A homology model of Salmonella FliMM was built based on the crystal structure of Thermotoga maritima FliMM (PDB code 3SOH). (C) Crystal structure of Salmonella FliMC-FliNN fusion protein (PDB code 4YXB) and location of the FliN(E95G) suppressor mutation. FliMC and FliNN subunits are shown by green and cyan Cα ribbon models, respectively.
