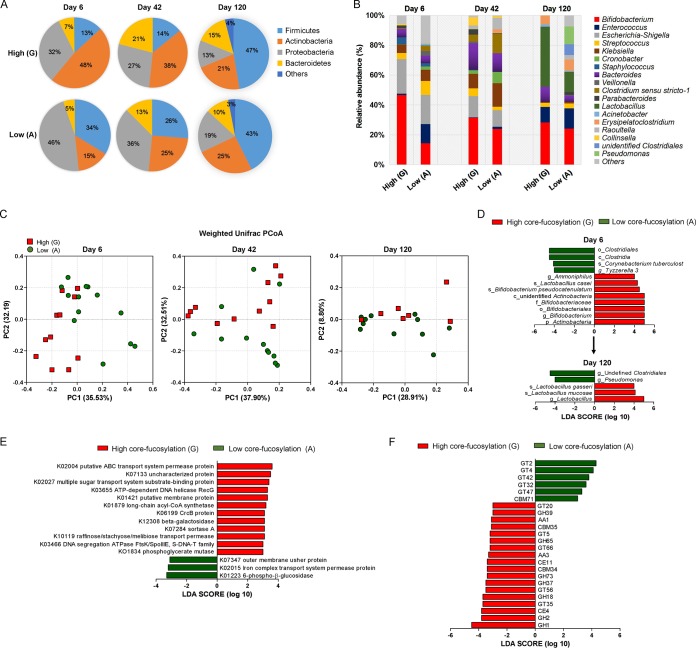
FIG 2.
Core fucosylation of milk N-glycan regulates the gut microbiome of infants. (A) The fecal microbiota profiles of infants fed by mothers with high (G group)- or low (A group)-core-fucosylated milk N-glycan at the phylum level based on 16S rRNA gene sequencing. (B) The fecal microbiota profiles of infants at genus level. (C) Characteristics of infant gut microbiota at days 6, 42, and 120 postpartum, illustrated by principal-coordinate analysis (PCoA) clustering analyses. Data from individuals (points) were clustered, and the centers of gravity were computed for each class. (D) The linear discriminant analysis effect size (LefSe) was adopted to identify the bacterial groups that showed significant differences in abundance between the high (G) and low (A) groups at day 6 postpartum. (E) Use of the Kyoto Encyclopedia of Genes and Genomes (KEGG) to evaluate the gut microbial functions between the high and low groups at day 42 postpartum. In order to screen the marker genes with significant differences between groups, the difference function between different groups was first detected by rank sum test and the dimension reduction was evaluated by LDA (linear discriminant analysis). The length of the histogram represents the influence of the difference function. (F) The Carbohydrate-Active enZYmes Database (CAZy) was used to evaluate the gut microbial functions of infants within the high and low groups at day 42 postpartum. GHs, glycoside hydrolases; GTs, glycosyl transferases; PLs, polysaccharide lyases; CEs, carbohydrate esterases; AAs, auxiliary activities; CBMs, carbohydrate-binding modules.