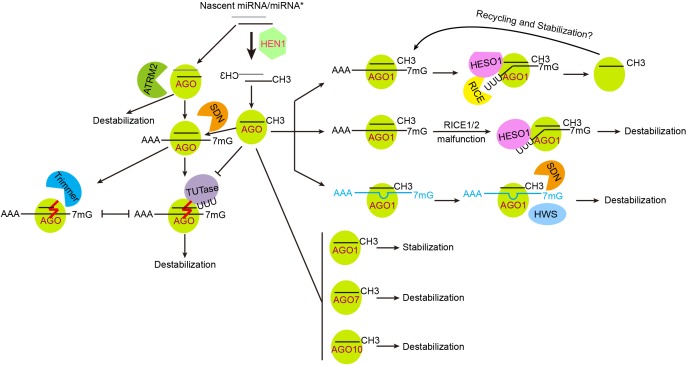
FIGURE 2.
Regulation of miRNA stability and turnover. Plant miRNAs are heavily methylated, which is crucial for their stabilization. ATRM2 is involved in the degradation of unmethylated miRNA/miRNA∗s, likely during the initiation of RISC loading. RISC-associated unmethylated miRNAs are destabilized via 3′ end tailing and 3′-to-5′ trimming. TUTase, terminal uridylyl transferase such as HESO1 and URT1 in Arabidopsis and MUT68 in Chlamydomonas reinhardtii. Trimmer, a yet uncharacterized enzyme catalyzing 3′-to-5′ trimming of unmethylated miRNAs. The effects of AGO proteins on miRNA stability are judged only by the results of ago mutants and/or over-expression transgenic plants, and are not necessarily absolute. Defects in the degradation of cleavage products (e.g., in the RICE1/2 malfunction backgrounds) cause miRNA reduction, suggesting that successful release of cleavage products may facilitate RISC recycling and stabilization. TM (in blue color) induced miRNA destabilization involves the actions of SDNs and an F-box protein HWS.