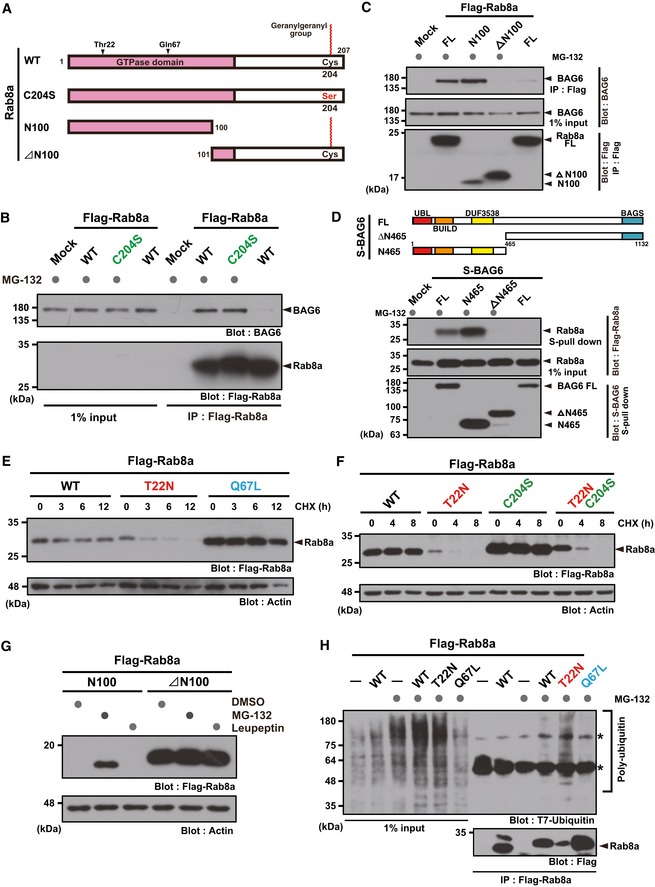
Figure 3. GDP‐bound form of the Rab8a GTPase domain is critical for its ubiquitin‐mediated degradation.
-
ASchematic representation of Rab8a protein. To prevent C‐terminal geranylgeranylation, the Cys204 residue was substituted with a serine residue. Alternatively, two truncated proteins (N100 and ∆N100 fragments of Rab8a) were prepared for this experiment.
-
BC‐terminal geranylgeranylation of Rab8a was dispensable for BAG6 recognition.
-
CThe N‐terminal GTPase domain of Rab8a was essential for BAG6 recognition. A series of Flag‐tagged truncated fragments of WT Rab8a were expressed in HeLa cells with S‐tagged BAG6 and treated with (+) or without (−) protease inhibitors for 4 h. Flag‐Rab8a substrates were immunoprecipitated and probed with an anti‐BAG6 antibody.
-
DHydrophobicity recognition domain of BAG6 (N465) was critical for Rab8a recognition. A schematic representation of the BAG6‐truncated proteins used in this experiment is shown in the upper panel. Numbers denote the corresponding amino acids of mammalian BAG6. Positions of UBL, BUILD, and DUF3538 domains, which are all linked to hydrophobicity recognition by BAG6 37, are indicated. WT, wild‐type; ΔN465, N‐terminal 465 residues‐deleted mutant; and Ν465, C‐terminal 689 residues‐deleted mutant.
-
E, FCHX chase experiments show that GDP‐bound Rab8a (T22N) was a highly labile protein, while the GTP‐bound active mutant (Q67L) was a stable protein (E). Instability of the T22N mutant was not perturbed by the C204S mutation (F). Actin was used as a loading control.
-
GN‐terminal 100 residue fragment of the Rab8a GTPase domain was sensitive to the proteasome inhibitor, while the ∆N100 fragment was not.
-
HRab8a (T22N) was polyubiquitinated in the presence of the proteasome inhibitor. Asterisks indicate non‐specific bands.
Source data are available online for this figure.
