Figure 4. The Switch I region of Rab8a is critical for BAG6 recognition and instability.
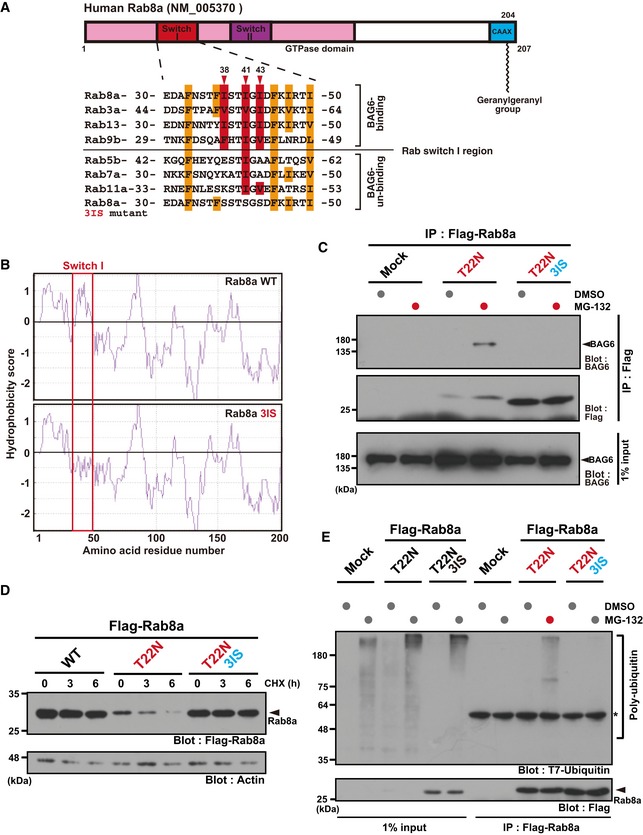
- Schematic representation of the two Switch regions (I and II) within the Rab8a GTPase domain. Numbers denote the corresponding amino acids of human Rab8a (upper panel). Amino acid sequence alignments of the Switch I region of Rab family proteins (lower panel). Three conserved hydrophobic residues (Ile38, Ile41, and Ile43) in this region are indicated in red, while the other hydrophobic residues are indicated in orange. The three hydrophobic residues were substituted with serine and this construct was designated as the T22N‐3IS mutant. Note that Ile38 and Ile43 of Rab8a are not conserved in Rab7.
- Kyte‐Doolittle hydrophobicity plots of the complete amino acid sequence of human WT Rab8a and T22N‐3IS mutant. The hydrophobicity peak within WT Switch I was abolished in the 3IS mutant (indicated within the red box). The numbers on the horizontal axis denote the corresponding amino acid positions in these proteins.
- Substitution of the hydrophobic residues of Switch I with hydrophilic residues (T22N 3IS) abolished its binding to BAG6 protein.
- Rab8a (T22N‐3IS) mutant protein was highly stable, while Rab8a (T22N) protein was quite unstable in HeLa cells. Actin was used as a loading control.
- Rab8a (T22N‐3IS) mutant was not subject to polyubiquitin co‐precipitation, while Rab8a (T22N) protein was.
Source data are available online for this figure.
