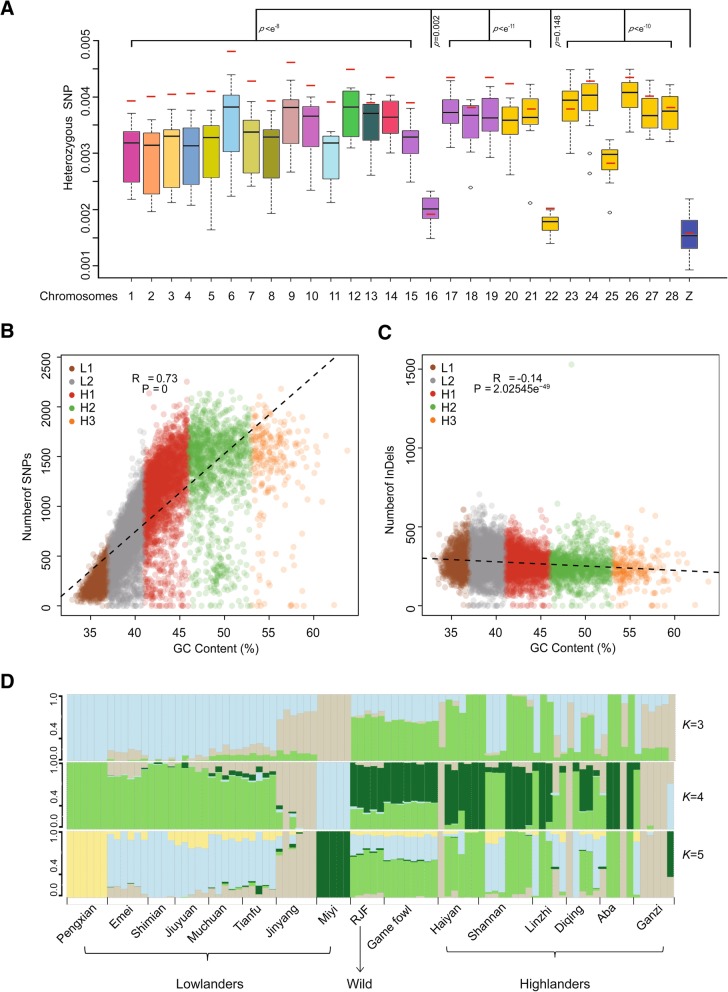
Fig. 1.
Heterozygous SNPs in each chicken chromosome; correlation between variations and GC content of the genome and population structures. a Boxplot showing the difference in the number of heterozygous SNP per base pair between the Z chromosome and all autosomes. The black lines inside the boxes represent the median of all 86 domestic chickens. The red lines represent the median of all five RJFs. A two-tailed t-test was conducted to detect significant differences between the Z chromosome and all autosomes. Correlation between the number of SNPs (b) or the number of indels (c) in 91 chickens and the GC level in the isochores of the chicken genome. d Population structures with the number of ancestral clusters, K, ranging from 3 to 5