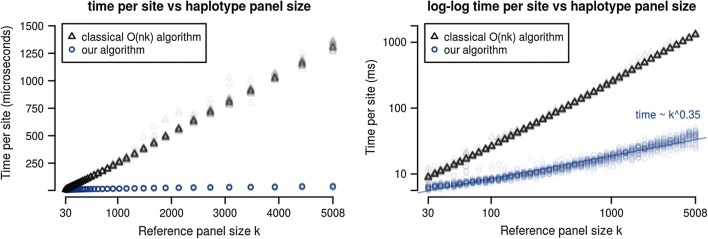
Fig. 7.
Runtime per site for conventional linear algorithm vs our sparse-lazy algorithm. Runtime per site as a function of haplotype reference panel size k for our algorithm (blue) as compared to the classical linear time algorithm (black). Both were implemented in C++ and benchmarked using datasets preloaded into memory. Forward probabilities are calculated for randomly generated haplotypes simulated by a recombination–mutation process, against random subsets of the 1000 genomes dataset