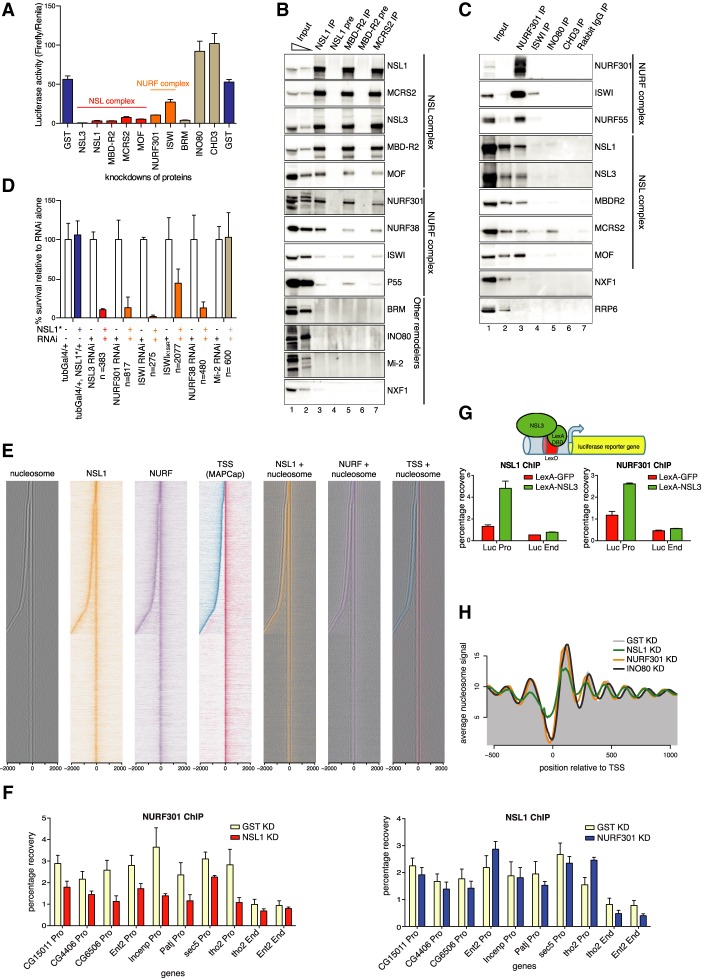
Figure 2.
The NSL complex recruits the NURF complex and maintains nucleosome pattern at promoters. (A) Luciferase activity (ratio of Firefly luciferase/Renila luciferase) in a Gal4-NSL3 reporter system in knockdowns for GST (control, far left and right), NSL and NURF complex members, BRM, INO80, and CHD3. Error bars, SD of three independent experiments. (B) Immunoprecipitation of the endogenous NSL complex members NSL1, MBDR2, and MCRS2. Preimmune Sera (NSL1, MBD-R2) served as controls. The western blot was probed by antibodies against the NSL and NURF complexes, other chromatin remodelers, and NXF1, a protein involved in mRNA export (as a negative control). (C) Immunoprecipitation of the chromatin remodelers NURF301, ISWI, INO80, and CHD3. Rabbit IgG served as a control. The western blot was probed by antibodies against the NSL and NURF complexes. INO80 immunoprecipitates MCRS2, but not other NSL complex members. NXF1 and RRP6 served as negative controls. (D) Bar chart showing of viability of flies when combining knockdown of NSL3, NURF301, ISWI (dominant-negative mutant, marked with asterisk, and RNAi), NURF38, or Mi-2 with heterozygous NSL1 mutant (blue bars) relative to RNAi/mutant alone (white bars). Heterozygous NSL1 mutant alone (blue bar) does not cause lethality. The mean ± SD of at least three independent crosses is shown. The numbers of flies counted are indicated by n. (E) Heat maps displaying the input-normalized ChIP enrichments of NSL1 (orange) and NURF301 (purple) ±2 kb around the MAPCap TSS. Nucleosome signal is depicted in black. MAPCap TSS positions are indicated in red (Watson strand) and blue (Crick strand). (Right) Nucleosome signals are overlaid with the ChIP combined with high-throughput sequencing (ChIP-seq) and TSS signal. (F) ChIP-qPCR analyses for NURF301 (top) and NSL1 (bottom) for knockdowns against GST (pale yellow), NSL1 (red), and NURF301 (blue). qPCR analysis was performed with primer sets positioned at the promoters (Pro) and ends (End) of indicated genes. Results are expressed as mean (±SD) of relative percentage recovery of immunoprecipitated material over input material. (G) Schematic: LexA-NSL3 is used to activate expression of lexO-luciferase reporter in transgenic flies. Bar charts: ChIP-qPCR experiments performed with NSL1 (left) and NURF301 (right) antibodies and primer pairs specific for promoter and end regions of the reporter gene. Results are expressed as mean (±SD) of the relative percentage recovery of immunoprecipitated material over input material. (H) Summary plot depicting the nucleosome signal −500 to 1000 bp of the TSS in wild-type (gray shaded area), NSL1-depleted (green), NURF301-depleted (orange), and INO80-depleted (purple) cells.