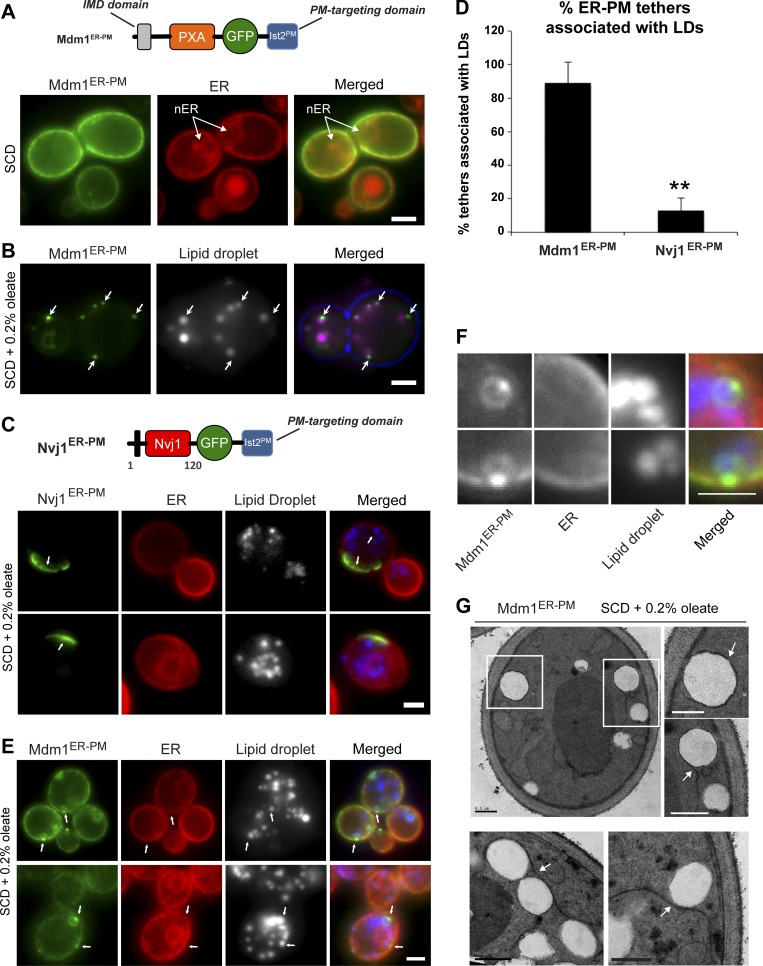
Figure 2.
N-terminal region of Mdm1 is sufficient to define sites of LD budding. (A) Top: Diagram of GFP-tagged Mdm1ER-PM chimera with a C-terminal Ist2 PM-targeting domain. Bottom: Light microscopy of yeast expressing GFP-tagged Mdm1ER-PM chimera with Ds-Red HDEL marker to label the ER. Scale bar, 2 µm. (B) Light microscopy of yeast fed 0.2% oleate and expressing GFP-tagged Mdm1ER-PM chimera. LDs (gray) visualized by MDH staining (arrows). Scale bar, 2 µm. (C) Top: Diagram of GFP-tagged Nvj1ER-PM chimera with a C-terminal Ist2 PM-targeting domain. Bottom: Light microscopy of yeast fed 0.2% oleate and expressing GFP-tagged Nvj1ER-PM chimera with Ds-Red HDEL marker to label the ER. LDs (gray) visualized by MDH staining (arrows). Scale bar, 2 µm. (D) Quantification of yeast ER–PM tethers associated with LDs. Percentage of yeast cells over the total number of cells counted; mean ± SD; n > 50 cells; **, P < 0.005; Student’s t test. (E) Light microscopy of yeast fed 0.2% oleate and expressing GFP-tagged Mdm1ER-PM chimera with Ds-Red HDEL marker to label the ER. LDs (gray) visualized by MDH staining (arrows). Scale bar, 2 µm. (F) Higher magnification of images in E. Scale bar, 2 µm. (G) Thin-sectioning TEM of Mdm1ER-PM fed 0.2% oleate. Arrows indicate ER wrapping. Scale bars, 0.5 µm.