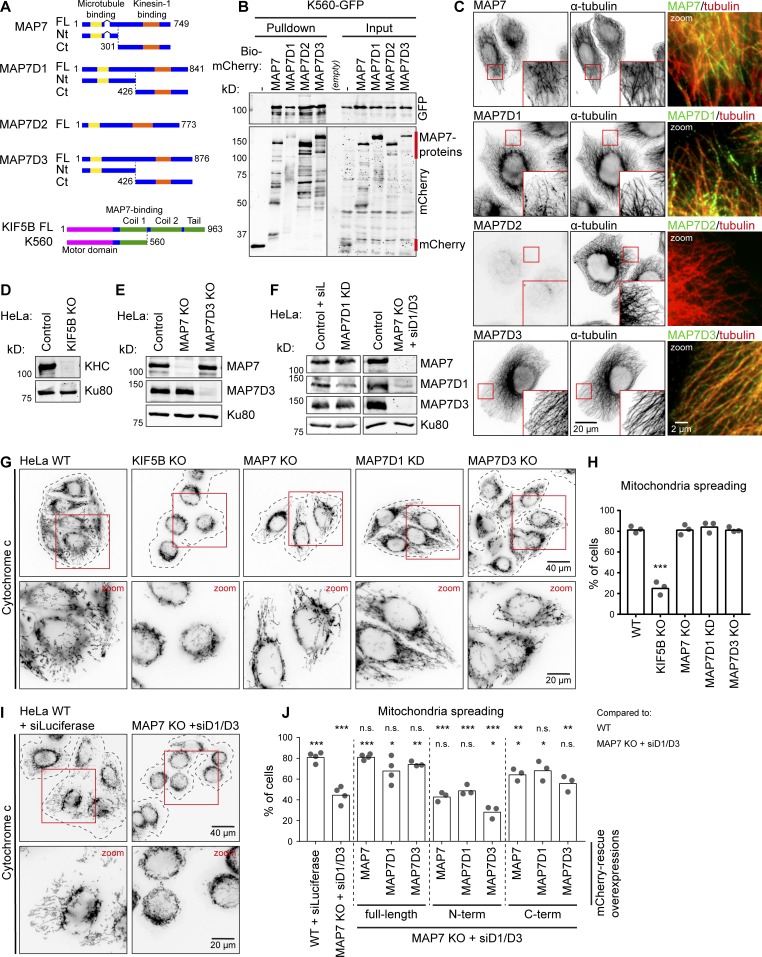
Figure 1.
Redundant function of MAP7 family proteins in kinesin-1–dependent mitochondria distribution. (A) Schemes of MAP7 family proteins and KIF5B constructs. (B) Streptavidin pull-down assay with extracts of HEK293T cells expressing BirA, K560-GFP (prey), and the indicated Bio-mCherry–labeled proteins (bait) analyzed by Western blotting. Red lines indicate the position of mCherry (negative control) and MAP7 proteins. (C) Immunostaining of HeLa cells for endogenous MAP7 family members and α-tubulin imaged on a widefield microscope. (D–F) Western blot analysis of the indicated HeLa knockout (KO) and knockdown (KD) cells with the indicated antibodies; Ku80 was used as a loading control. (G and I) HeLa cells treated as indicated stained for mitochondria (cytochrome c). Cell outlines are indicated with gray dashed lines; zooms (red squares) are shown below. (H and J) Mitochondria distribution scored per condition using cytochrome c staining. (H) n = 345, 347, 332, 338, and 373 cells from three independent experiments; WT vs KIF5B KO, P = 0.0002, t test. (J) n = 444 (WT + siLuciferase), n = 589 (MAP7 KO + siMAP7D1/D3), and for rescue conditions on top of MAP7 KO + siMAP7D1/D3: n = 261, 277, 324, 297, 277, 296, 263, 267, and 416 cells all from three or four independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; t test. Ct, C terminal; FL, full length; Nt, N terminus.