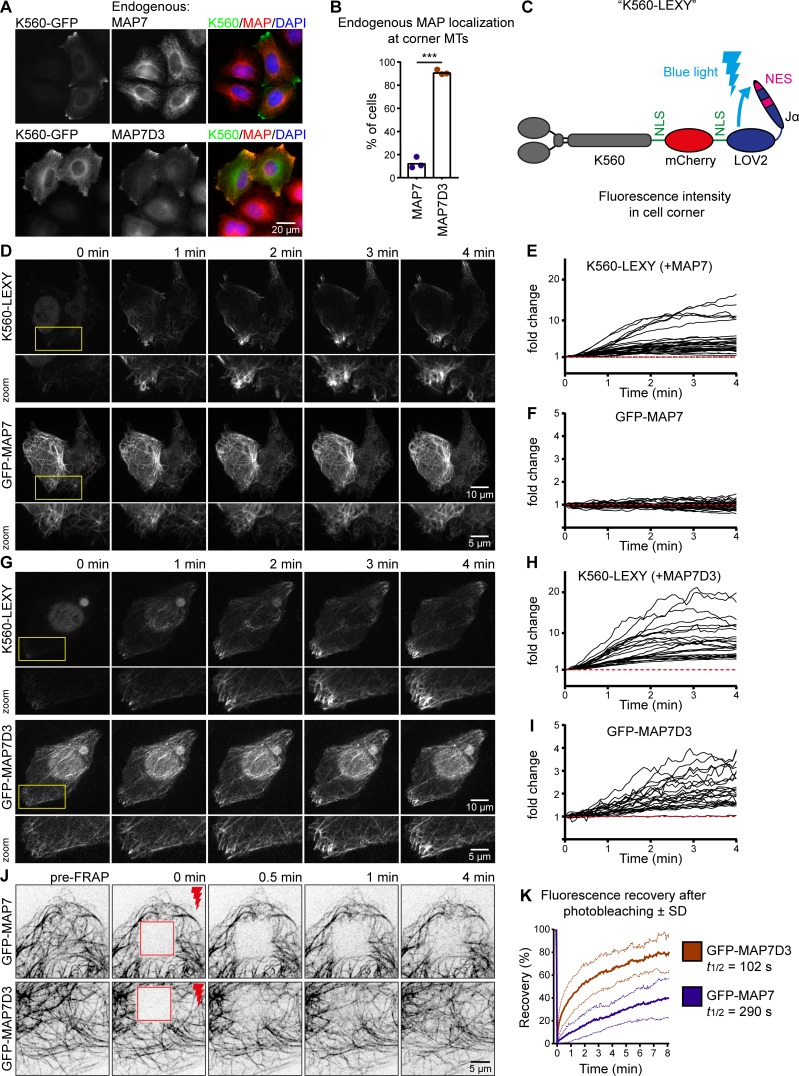
Figure 4.
Kinesin-1 can redistribute MAP7D3 in cells. (A and B) Widefield images of K560-GFP overexpressed in HeLa cells stained for endogenous MAP7 or MAP7D3 (A) used to quantify endogenous MAP localization (B). n = 232 and 303 cells from three independent experiments, P < 0.001, t test. (C) Scheme of the K560-LEXY construct containing two NLS sequences and an mCherry tag. Blue light induces a conformation change, causing detachment of the Jα-peptide containing a nuclear export signal from the LOV2 domain. (D and G) Single frames of KIF5B KO cells cotransfected with K560-LEXY and GFP-MAP7 (D) or GFP-MAP7D3 (G) sequentially illuminated with green and blue light (in that specific order). Zooms are indicated in yellow. (E, F, H, and I) Measurements of fluorescence intensity changes over time in K560-LEXY–positive cell corners. Black lines represent single measurements of K560-LEXY (E and H), GFP-MAP7 (F), and GFP-MAP7D3 (I). n = 31 measurements from 17 cells (E and F) and n = 22 measurements from 14 cells (H and I) from two independent experiments. (J) Single frames of FRAP experiments on COS7 cells overexpressing GFP-MAP7 or -MAP7D3. Stills show a baseline (pre-FRAP), the first frame after photobleaching (0 min) and the indicated time points after FRAP of a 10 × 10-µm-square region (shown in red). (K) Quantification of fluorescence recovery of J. The graph shows mean curves (bold lines) ± SD (light dotted lines) over time. n = 18 cells from three independent experiments for each condition.