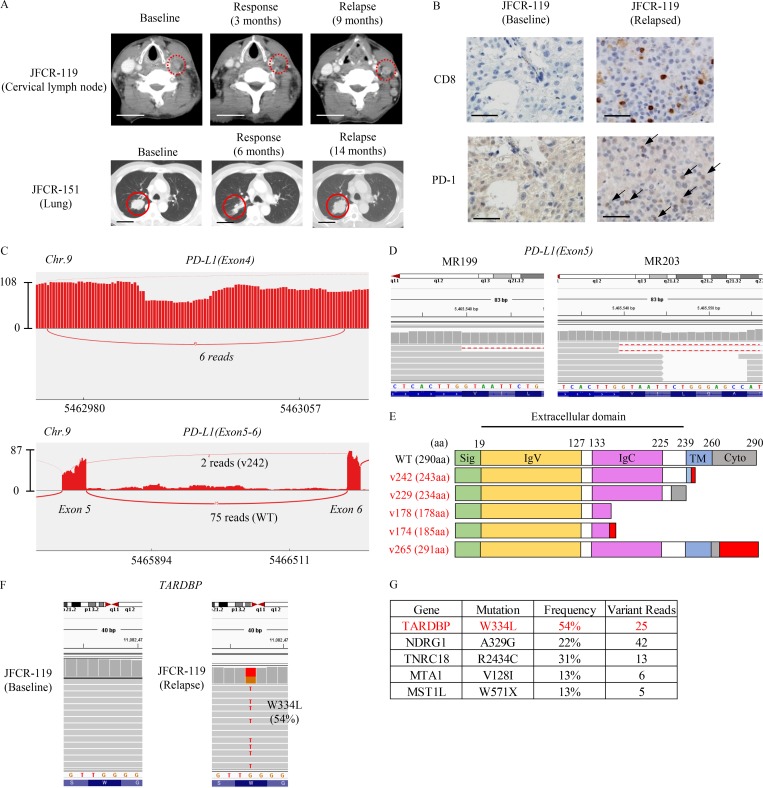
Figure 1.
Identification of PD-L1 C-terminal–deficient splicing variants in patients who were relapsed from PD-L1 blockade therapy. (A) Representative computed tomographic images of JFCR-119 and JFCR-151 at baseline and at the time of relapse. Bars, 5 cm. (B) Representative IHC staining of CD8 and PD-1 at baseline and in the relapsed tumor from JFCR-119. Bars, 10 µm. (C) Sashimi plot RNA-seq analysis of the PD-L1 spliced region. The figure shows representative data for PD-L1v178 (above) and PD-L1v242 (below) in MR203. (D) Integrative genomics viewer (IGV) data indicating PD-L1v242 in MR199 and MR203 are shown. (E) The PD-L1 splicing variants identified from JFCR-119 and JFCR-151. The domains are indicated as follows: signal peptide (Sig) on 1–18 aa as green; IgV domain on 19–127 aa as yellow; IgC domain on 133–225 aa as pink; transmembrane domain (TM) on 239–259 aa as blue; and cytoplasmic domain (Cyto) on 260–290 aa as gray. The red region demonstrates the additional amino acids from aberrant splicing. (F) IGV data of TARDBP on the mutated region in pretreatment and relapsed samples of JFCR-119. (G) Relapsed tumor-specific mutations in JFCR-119 analyzed by RNA-seq.